You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004640_00568
You are here: Home > Sequence: MGYG000004640_00568
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
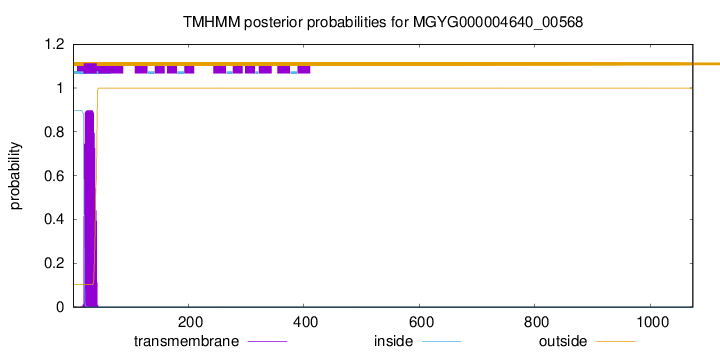
TMHMM annotations
Basic Information help
| Species | Prevotella sp900551055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900551055 | |||||||||||
| CAZyme ID | MGYG000004640_00568 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2341; End: 5565 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 39 | 710 | 3.3e-101 | 0.6409574468085106 |
| CBM57 | 926 | 1068 | 4.8e-25 | 0.9727891156462585 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.05e-50 | 89 | 523 | 31 | 427 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam18368 | Ig_GlcNase | 2.69e-35 | 778 | 888 | 1 | 104 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| pfam11721 | Malectin | 2.33e-25 | 926 | 1069 | 4 | 164 | Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognizes and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. |
| PRK10340 | ebgA | 6.37e-23 | 136 | 526 | 113 | 475 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 1.58e-22 | 133 | 523 | 66 | 443 | beta-D-glucuronidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANI89980.1 | 0.0 | 34 | 911 | 17 | 876 |
| AEU36287.1 | 3.29e-291 | 50 | 907 | 29 | 868 |
| ADV83491.1 | 3.33e-290 | 22 | 907 | 26 | 890 |
| ADW69333.1 | 1.63e-275 | 17 | 907 | 13 | 889 |
| QND52244.1 | 1.01e-197 | 67 | 888 | 76 | 921 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VZO_A | 6.92e-81 | 82 | 887 | 71 | 890 | Crystalstructure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZS_A | 1.75e-80 | 82 | 887 | 71 | 890 | ChitosanProduct complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
| 2VZT_A | 1.11e-79 | 82 | 887 | 71 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
| 2VZU_A | 5.16e-79 | 82 | 887 | 71 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
| 5N6U_A | 5.53e-58 | 86 | 889 | 37 | 831 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82NR8 | 7.25e-104 | 66 | 887 | 56 | 895 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
| Q5H7P5 | 4.92e-95 | 64 | 887 | 7 | 944 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
| Q75W54 | 6.99e-93 | 64 | 887 | 10 | 935 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| Q56F26 | 9.56e-80 | 82 | 887 | 71 | 890 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
| D4AUH1 | 1.34e-68 | 43 | 889 | 34 | 878 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
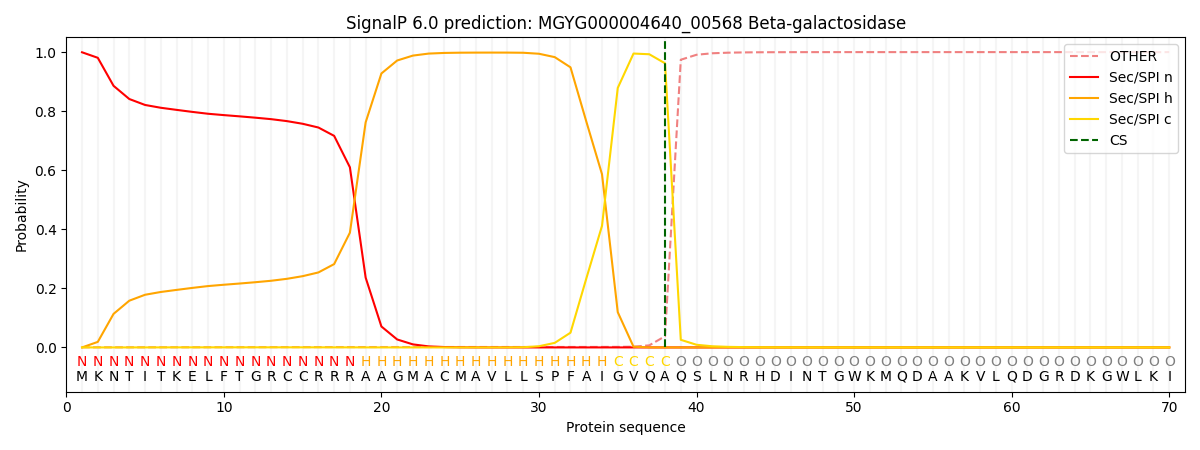
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001171 | 0.997832 | 0.000290 | 0.000317 | 0.000195 | 0.000166 |