You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004650_01548
You are here: Home > Sequence: MGYG000004650_01548
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Odoribacter; | |||||||||||
| CAZyme ID | MGYG000004650_01548 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1802; End: 3343 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 182 | 490 | 6.8e-49 | 0.8212290502793296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4833 | COG4833 | 2.96e-16 | 196 | 444 | 45 | 285 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| cd04791 | LanC_SerThrkinase | 0.007 | 260 | 339 | 91 | 165 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
| cd04792 | LanM-like | 0.007 | 260 | 363 | 656 | 762 | Cyclases involved in the biosynthesis of class II lantibiotics, and similar proteins. LanM-like proteins. LanM is a bifunctional enzyme, involved in the synthesis of class II lantibiotics. It is responsible for both the dehydration and the cyclization of the precursor-peptide during lantibiotic synthesis. The C-terminal domain shows similarity to LanC, the cyclase component of the lan operon, but the N terminus seems to be unrelated to the dehydratase, LanB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26770.1 | 0.0 | 1 | 513 | 1 | 513 |
| QUR45037.1 | 0.0 | 1 | 513 | 1 | 513 |
| BCA48077.1 | 5.56e-186 | 21 | 506 | 3 | 491 |
| ALJ44322.1 | 5.56e-186 | 23 | 506 | 5 | 491 |
| QQA08375.1 | 5.56e-186 | 23 | 506 | 5 | 491 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SHD_A | 9.60e-26 | 175 | 501 | 72 | 384 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 2.04e-24 | 175 | 501 | 73 | 385 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 1.25e-23 | 175 | 501 | 73 | 385 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
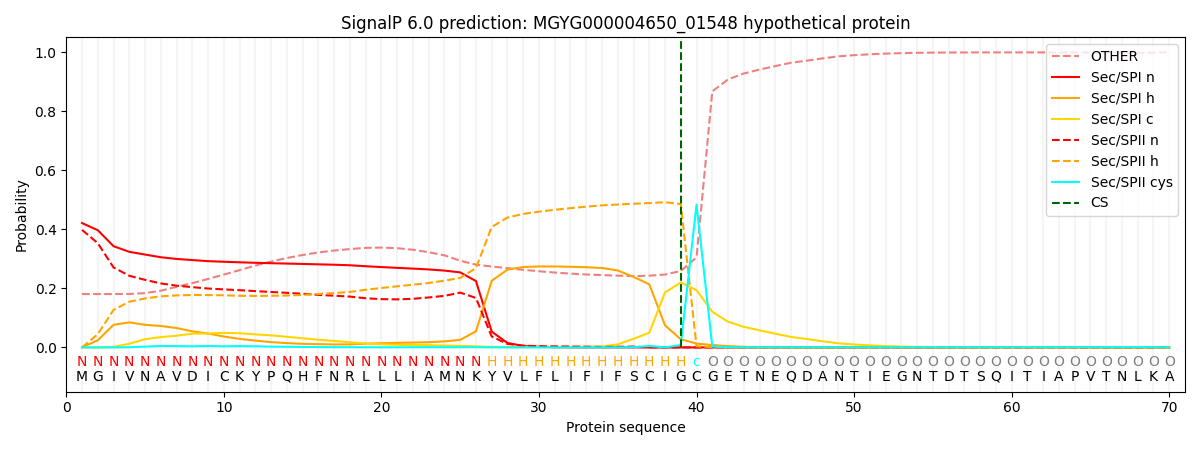
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.184091 | 0.303751 | 0.511210 | 0.000152 | 0.000524 | 0.000277 |
