You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004658_02321
You are here: Home > Sequence: MGYG000004658_02321
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes finegoldii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes finegoldii | |||||||||||
| CAZyme ID | MGYG000004658_02321 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39325; End: 40536 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 56 | 397 | 1.9e-72 | 0.8910614525139665 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 1.31e-35 | 78 | 323 | 31 | 287 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 5.21e-31 | 44 | 351 | 3 | 301 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam07221 | GlcNAc_2-epim | 6.87e-05 | 153 | 301 | 153 | 314 | N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase). This family contains a number of eukaryotic and bacterial N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase) enzymes (EC:5.3.1.8) approximately 500 residues long. This converts N-acyl-D-glucosamine to N-acyl-D-mannosamine. |
| cd00249 | AGE | 1.04e-04 | 152 | 301 | 185 | 340 | AGE domain; N-acyl-D-glucosamine 2-epimerase domain; Responsible for intermediate epimerization during biosynthesis of N-acetylneuraminic acid. Catalytic mechanism is believed to be via nucleotide elimination and readdition and is ATP modulated. AGE is structurally and mechanistically distinct from the other four types of epimerases. The AGE domain monomer is composed of an alpha(6)/alpha(6)-barrel, the structure of which is also found in glucoamylase and cellulase. The active form is a homodimer. The alignment also contains subtype III mannose 6-phosphate isomerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFL76820.1 | 3.89e-309 | 1 | 403 | 1 | 403 |
| QRQ62402.1 | 7.09e-80 | 1 | 399 | 1 | 367 |
| APU98500.1 | 1.58e-78 | 1 | 399 | 1 | 367 |
| VTR42877.1 | 1.39e-76 | 1 | 399 | 1 | 367 |
| QIH35643.1 | 3.07e-75 | 1 | 399 | 1 | 367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SHD_A | 1.42e-55 | 43 | 401 | 53 | 389 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 4.28e-54 | 43 | 401 | 54 | 390 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 3.24e-53 | 43 | 401 | 54 | 390 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 4D4A_A | 7.57e-34 | 38 | 356 | 21 | 325 | ChainA, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
| 4BOK_A | 9.72e-34 | 49 | 356 | 8 | 304 | ChainA, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
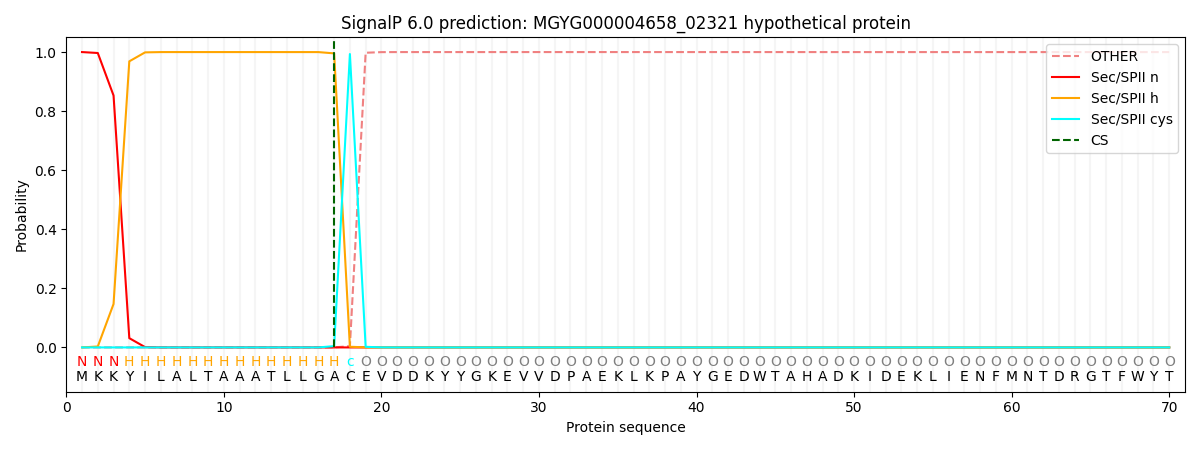
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000068 | 0.000000 | 0.000000 | 0.000000 |
