You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004668_00001
You are here: Home > Sequence: MGYG000004668_00001
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
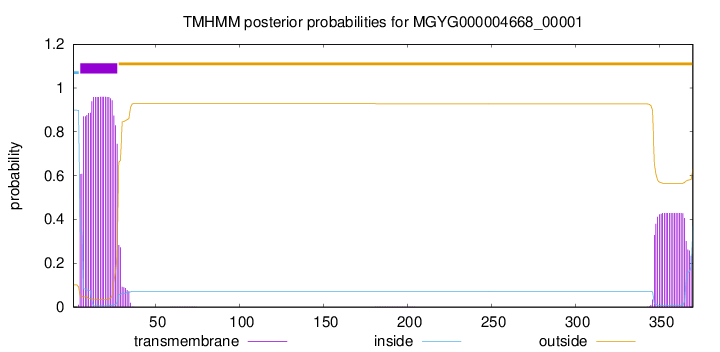
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Limosilactobacillus; | |||||||||||
| CAZyme ID | MGYG000004668_00001 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 113; End: 1225 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 2.05e-55 | 159 | 316 | 39 | 194 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| PRK08581 | PRK08581 | 1.48e-45 | 166 | 310 | 323 | 472 | amidase domain-containing protein. |
| PRK06347 | PRK06347 | 8.35e-38 | 125 | 316 | 98 | 309 | 1,4-beta-N-acetylmuramoylhydrolase. |
| smart00047 | LYZ2 | 5.24e-32 | 164 | 310 | 9 | 147 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| PRK05684 | flgJ | 1.17e-23 | 114 | 301 | 100 | 294 | flagellar assembly peptidoglycan hydrolase FlgJ. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZN92542.1 | 6.82e-121 | 1 | 368 | 1 | 360 |
| QFV01117.1 | 3.00e-92 | 1 | 369 | 3 | 380 |
| QFS34532.1 | 8.81e-75 | 10 | 369 | 10 | 349 |
| QFG72894.1 | 1.35e-69 | 150 | 369 | 2 | 221 |
| AEI56515.1 | 1.78e-67 | 10 | 370 | 13 | 343 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3FI7_A | 1.16e-34 | 163 | 310 | 30 | 183 | CrystalStructure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain [Listeria monocytogenes EGD-e] |
| 5T1Q_A | 9.72e-32 | 162 | 310 | 59 | 212 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 5DN5_A | 2.02e-15 | 162 | 301 | 2 | 145 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5DN4_A | 2.80e-15 | 162 | 301 | 2 | 145 | Structureof the glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 3VWO_A | 2.30e-13 | 171 | 302 | 13 | 144 | Crystalstructure of peptidoglycan hydrolase mutant from Sphingomonas sp. A1 [Sphingomonas sp. A1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9CIT4 | 1.57e-34 | 157 | 323 | 56 | 227 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. lactis (strain IL1403) OX=272623 GN=acmA PE=3 SV=1 |
| A2RHZ5 | 1.06e-33 | 157 | 316 | 56 | 220 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. cremoris (strain MG1363) OX=416870 GN=acmA PE=3 SV=1 |
| P0C2T5 | 7.41e-33 | 157 | 316 | 56 | 220 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. cremoris OX=1359 GN=acmA PE=3 SV=1 |
| Q2G222 | 4.23e-30 | 162 | 310 | 319 | 472 | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=SAOUHSC_02979 PE=1 SV=1 |
| P37710 | 4.29e-30 | 159 | 310 | 174 | 333 | Autolysin OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0799 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
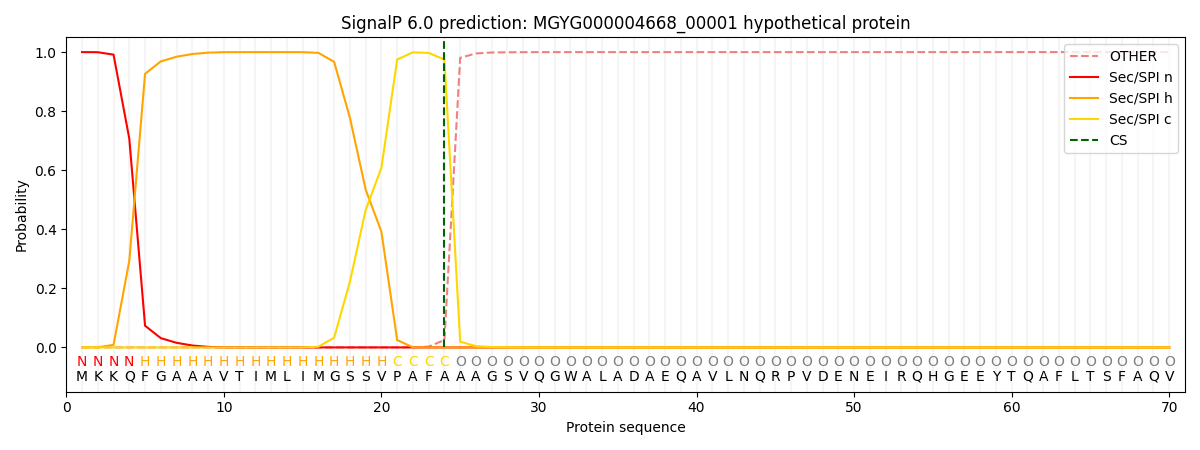
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.998963 | 0.000172 | 0.000214 | 0.000173 | 0.000156 |