You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004676_00169
You are here: Home > Sequence: MGYG000004676_00169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766005 | |||||||||||
| CAZyme ID | MGYG000004676_00169 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17408; End: 19936 Strand: - | |||||||||||
Full Sequence Download help
| MKNSFIKKSV CALGACCFCV GMAHADEWIR INQLGYLPQS VKVAVFMSDE ATSVQEYALV | 60 |
| DAFTKQTVKT FRAPQSTGAW ENMKSTYRLD FSDFVQPGTY YLKAGNAVSP TFPIAADVYK | 120 |
| GTADFLLNYL RQQRCGDNPR FDKKCHQKDA YIVYHPTKNG QHIDVRGGWH DASDYLQYTT | 180 |
| TTANAIYQMM FAYQQNPEAF GDEYGTDGRK GANGIPDIID EIRWGMDWLN RMNPEPGEFY | 240 |
| NQLADDRDHV SMRLPFLDPA DYGYGPGNGR PVYFCSGEKQ KRGKFMNETT GVASTTGKFA | 300 |
| SCFSLGSRIL KDFYPELAEQ IGKKAKDAFD MGVKKPGVCQ TASIVSPYIY EEDNWTDDME | 360 |
| LAAMELYHAS GEKEYLNRAV EYARREPVTP WMGADSARHY QWYPFVNLGH YHLAKVKGNS | 420 |
| RLNQEFVRNL RAGIERTFEK AVKSPFLHGI PYIWCSNNLT VAMLTQCGLY RELTGDRKYA | 480 |
| EMEAAMLDWL LGCNPWGTSM IIELPLHGDY PSESHAALLL HKGYGQPTGG LVDGPVYNSI | 540 |
| FKGLRGVNLN GLPGLPGEKY EAVQPRTMVY HDVVNDYSTN EPTMDGTASL TYYLSLMEVE | 600 |
| GMKQGQLSKD LNRYVDGGIV GTNPGKKVIS LVFTSDDKVD GEEKIIRVLD KNKVKGSFFF | 660 |
| TGNFFRRYPE VVKRLLEKQH YVGSHSDKHL LYMPWENRDS LLVSRAEFEN DLLNSYREMS | 720 |
| KLGINYQDAP LFMPPYEYYN RTIAAWTKGL GLRVVNYTPG TLSNADYTTP DMGKSYRSSK | 780 |
| EIFEHLLAME KKEGLNGAIL LVHFGTDPKR TDKFYDTYLN KLIQTLKRKG YQFQTISEAV | 840 |
| AK | 842 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 119 | 594 | 1.4e-77 | 0.992822966507177 |
| CE4 | 631 | 756 | 3.5e-16 | 0.8846153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 6.32e-62 | 127 | 594 | 7 | 374 | Glycosyl hydrolase family 9. |
| cd10917 | CE4_NodB_like_6s_7s | 4.27e-30 | 627 | 827 | 1 | 171 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| cd02850 | E_set_Cellulase_N | 6.60e-28 | 28 | 114 | 1 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| pfam02927 | CelD_N | 4.60e-27 | 28 | 109 | 2 | 83 | Cellulase N-terminal ig-like domain. |
| cd10949 | CE4_BsPdaB_like | 2.17e-21 | 626 | 842 | 3 | 192 | Putative catalytic NodB homology domain of Bacillus subtilis putative polysaccharide deacetylase PdaB, and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by the putative polysaccharide deacetylase PdaB encoded by the pdaB gene on sporulation of Bacillus subtilis. Although its biochemical properties remain to be determined, the PdaB (YbaN) protein is essential for maintaining spores after the late stage of sporulation and is highly conserved in spore-forming bacteria. The glycans of the spore cortex may be candidate PdaB substrates. Based on sequence similarity, the family members are classified as carbohydrate esterase 4 (CE4) superfamily members. However, the classical His-His-Asp zinc-binding motif of CE4 esterases is missing in this family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT35459.1 | 0.0 | 25 | 840 | 21 | 834 |
| QUT66756.1 | 0.0 | 25 | 840 | 21 | 834 |
| QPH59586.1 | 0.0 | 25 | 840 | 21 | 834 |
| QUT98069.1 | 0.0 | 25 | 840 | 21 | 834 |
| QQA29127.1 | 0.0 | 25 | 840 | 21 | 834 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3X17_A | 1.59e-39 | 29 | 592 | 18 | 551 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 5U2O_A | 1.14e-28 | 37 | 595 | 2 | 537 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 5U0H_A | 4.66e-26 | 37 | 595 | 2 | 537 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 1UT9_A | 7.43e-26 | 29 | 598 | 7 | 605 | ChainA, CELLULOSE 1,4-BETA-CELLOBIOSIDASE [Acetivibrio thermocellus] |
| 1RQ5_A | 4.08e-25 | 29 | 534 | 7 | 552 | ChainA, Cellobiohydrolase [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14090 | 2.18e-23 | 29 | 601 | 341 | 911 | Endoglucanase C OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cenC PE=1 SV=2 |
| P0C2S1 | 2.39e-23 | 29 | 598 | 214 | 812 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus OX=1515 GN=celK PE=1 SV=1 |
| Q05156 | 2.48e-23 | 29 | 601 | 185 | 746 | Cellulase 1 OS=Streptomyces reticuli OX=1926 GN=cel1 PE=1 SV=1 |
| A3DCH1 | 5.47e-23 | 29 | 598 | 214 | 812 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
| P23658 | 1.49e-22 | 29 | 595 | 4 | 543 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
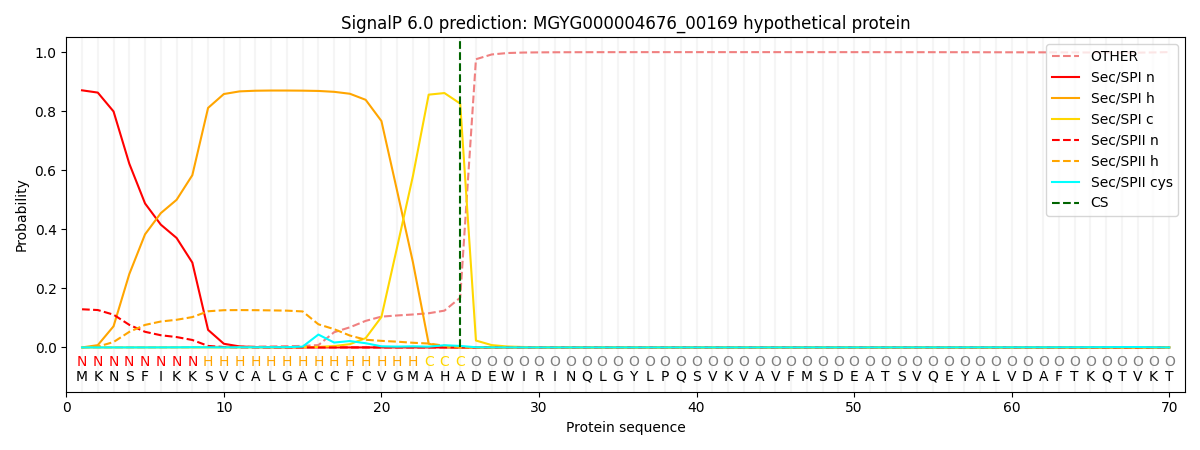
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001316 | 0.860524 | 0.137274 | 0.000307 | 0.000289 | 0.000262 |
