You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004676_00194
You are here: Home > Sequence: MGYG000004676_00194
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
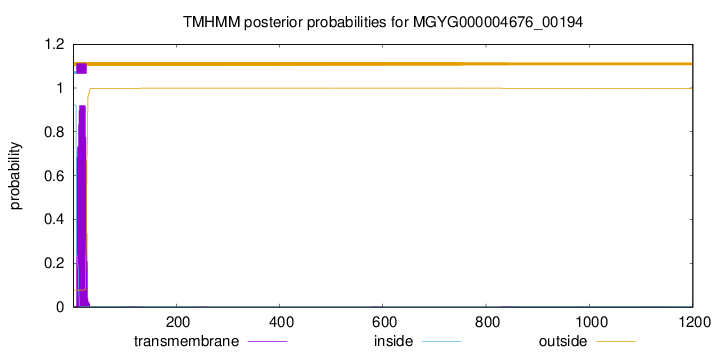
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900766005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900766005 | |||||||||||
| CAZyme ID | MGYG000004676_00194 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 60649; End: 64254 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH144 | 788 | 1199 | 4.6e-188 | 0.9900990099009901 |
| GH3 | 102 | 323 | 2.2e-72 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 0.0 | 26 | 775 | 26 | 764 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 2.78e-122 | 46 | 355 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 5.95e-106 | 45 | 445 | 1 | 376 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam10091 | Glycoamylase | 2.10e-100 | 968 | 1187 | 1 | 215 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
| COG5368 | COG5368 | 3.60e-93 | 784 | 1199 | 4 | 431 | Uncharacterized protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO67944.1 | 0.0 | 11 | 1199 | 12 | 1186 |
| QIU93909.1 | 0.0 | 6 | 775 | 1 | 770 |
| QMW85945.1 | 0.0 | 6 | 776 | 1 | 771 |
| AAO78673.1 | 0.0 | 6 | 776 | 1 | 771 |
| QQA08305.1 | 0.0 | 6 | 776 | 1 | 771 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5TF0_A | 0.0 | 26 | 776 | 1 | 751 | CrystalStructure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393],5TF0_B Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393] |
| 5XXL_A | 0.0 | 25 | 779 | 1 | 755 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 5XXN_A | 0.0 | 25 | 779 | 1 | 755 | CrystalStructure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXN_B Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXO_A Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482],5XXO_B Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482] |
| 4QT9_A | 6.32e-256 | 782 | 1199 | 13 | 430 | Crystalstructure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185],4QT9_B Crystal structure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185] |
| 3EU8_A | 1.11e-242 | 788 | 1201 | 16 | 429 | Crystalstructure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_B Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_C Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_D Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56078 | 2.69e-226 | 31 | 775 | 31 | 764 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| P33363 | 5.24e-223 | 31 | 775 | 31 | 764 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
| T2KMH0 | 2.77e-130 | 91 | 776 | 48 | 723 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| A6LGF6 | 2.99e-125 | 656 | 1196 | 184 | 717 | Exo beta-1,2-glucooligosaccharide sophorohydrolase (non-reducing end) OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=BDI_3064 PE=1 SV=1 |
| T2KMH9 | 1.59e-110 | 9 | 776 | 2 | 755 | Putative beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22230 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
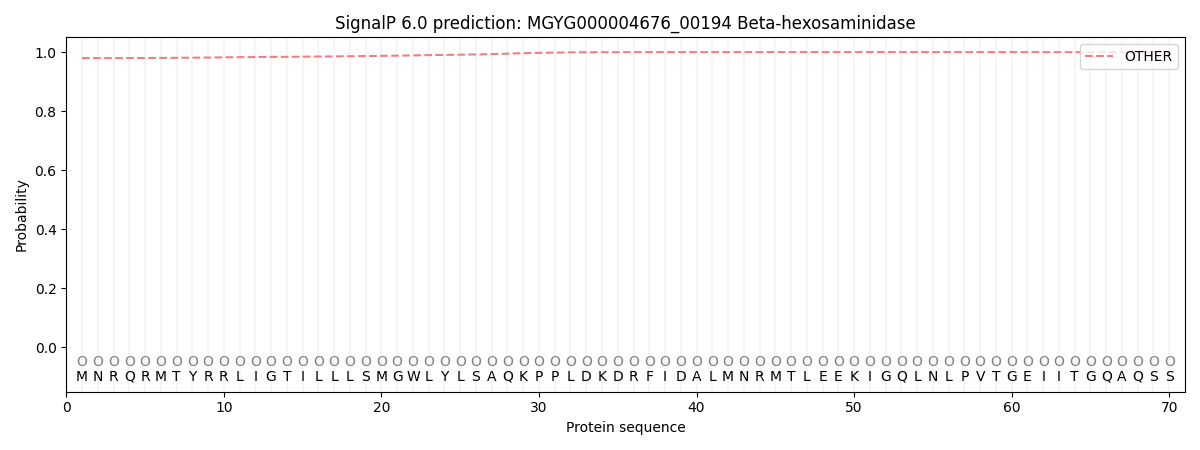
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.980029 | 0.007195 | 0.009539 | 0.000053 | 0.000033 | 0.003158 |