You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004688_01713
You are here: Home > Sequence: MGYG000004688_01713
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900550395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900550395 | |||||||||||
| CAZyme ID | MGYG000004688_01713 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30420; End: 32573 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 459 | 709 | 3.4e-45 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.95e-33 | 467 | 704 | 62 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.16e-32 | 467 | 706 | 104 | 307 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 8.16e-23 | 467 | 705 | 127 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 3.65e-07 | 58 | 139 | 6 | 85 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 0.010 | 82 | 140 | 51 | 109 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ48288.1 | 1.14e-252 | 14 | 711 | 14 | 716 |
| QUT46089.1 | 4.59e-252 | 14 | 711 | 14 | 716 |
| QIU93949.1 | 2.94e-217 | 1 | 715 | 1 | 729 |
| QDM11106.1 | 1.62e-216 | 1 | 717 | 1 | 731 |
| QGT73069.1 | 1.53e-214 | 1 | 715 | 1 | 729 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 1.61e-26 | 156 | 304 | 1 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 3.56e-22 | 165 | 302 | 4 | 142 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 1XYZ_A | 7.44e-17 | 467 | 714 | 132 | 343 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6WQW_A | 1.63e-15 | 456 | 704 | 99 | 323 | ChainA, Beta-xylanase [Thermobacillus composti] |
| 4W8L_A | 3.53e-15 | 453 | 704 | 100 | 338 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 2.27e-15 | 467 | 714 | 622 | 833 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| O69230 | 9.32e-14 | 453 | 704 | 466 | 704 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| Q96VB6 | 1.11e-13 | 467 | 711 | 130 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| Q60037 | 1.60e-13 | 460 | 704 | 472 | 686 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P07528 | 8.77e-13 | 457 | 704 | 162 | 391 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
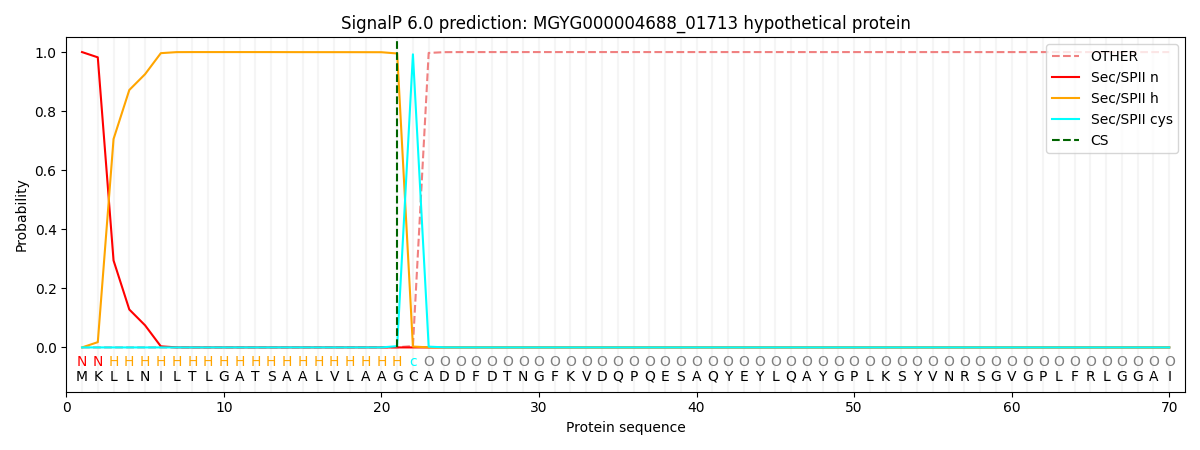
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000067 | 0.000000 | 0.000000 | 0.000000 |
