You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004693_01316
You are here: Home > Sequence: MGYG000004693_01316
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
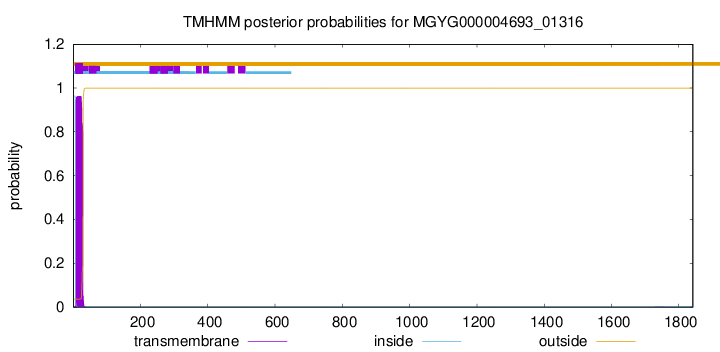
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UMGS1375; | |||||||||||
| CAZyme ID | MGYG000004693_01316 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6165; End: 11693 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 80 | 738 | 1.6e-139 | 0.6263297872340425 |
| GH2 | 887 | 1228 | 1.4e-51 | 0.3803191489361702 |
| CBM51 | 1453 | 1591 | 8.1e-27 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 1.58e-156 | 54 | 669 | 26 | 586 | beta-galactosidase. |
| PRK10340 | ebgA | 2.87e-146 | 45 | 669 | 4 | 560 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 9.22e-123 | 91 | 665 | 14 | 510 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02929 | Bgal_small_N | 4.33e-83 | 1080 | 1342 | 2 | 230 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| smart01038 | Bgal_small_N | 2.80e-77 | 1080 | 1342 | 2 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE90937.1 | 0.0 | 24 | 1590 | 20 | 1492 |
| CDI40488.1 | 0.0 | 24 | 1590 | 20 | 1492 |
| QTM99132.1 | 0.0 | 32 | 1591 | 23 | 1483 |
| AIQ34883.1 | 0.0 | 27 | 1596 | 19 | 1524 |
| APC49533.1 | 6.17e-277 | 22 | 1612 | 5 | 1304 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 1.60e-149 | 43 | 1328 | 3 | 966 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 1.64e-149 | 43 | 1328 | 4 | 967 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 9.40e-126 | 43 | 1345 | 6 | 997 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 2.08e-122 | 43 | 1334 | 9 | 991 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 1PX3_A | 4.83e-106 | 54 | 1338 | 25 | 1017 | ChainA, beta-galactosidase [Escherichia coli],1PX3_B Chain B, beta-galactosidase [Escherichia coli],1PX3_C Chain C, beta-galactosidase [Escherichia coli],1PX3_D Chain D, beta-galactosidase [Escherichia coli],1PX4_A Chain A, beta-galactosidase [Escherichia coli],1PX4_B Chain B, beta-galactosidase [Escherichia coli],1PX4_C Chain C, beta-galactosidase [Escherichia coli],1PX4_D Chain D, beta-galactosidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 7.39e-186 | 43 | 1345 | 19 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q56307 | 8.99e-149 | 43 | 1328 | 4 | 967 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q9K9C6 | 6.11e-140 | 44 | 1343 | 9 | 1012 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| A1SWB8 | 4.18e-122 | 53 | 1331 | 14 | 1022 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
| P24131 | 9.46e-119 | 43 | 1235 | 9 | 896 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
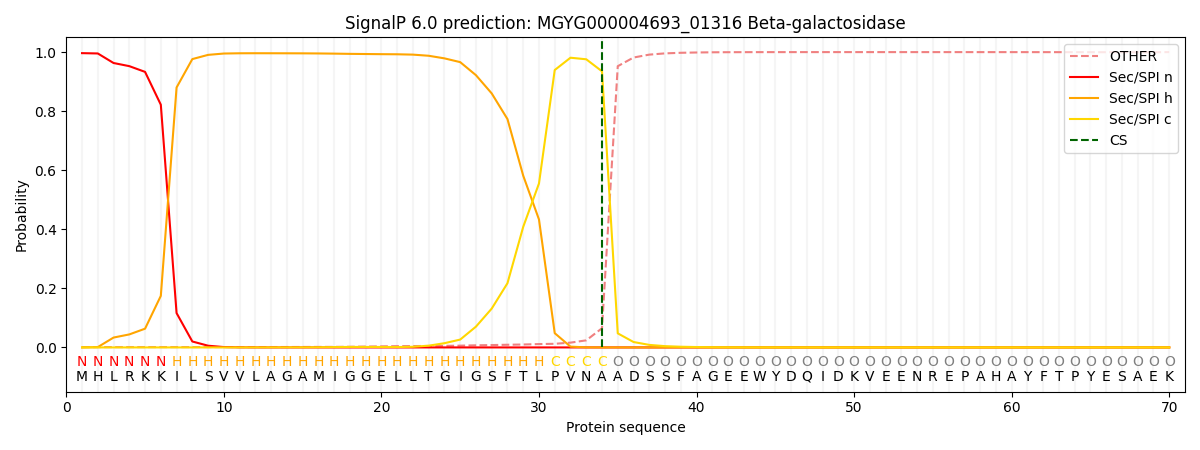
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002721 | 0.993484 | 0.002868 | 0.000456 | 0.000250 | 0.000206 |