You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004697_00651
You are here: Home > Sequence: MGYG000004697_00651
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
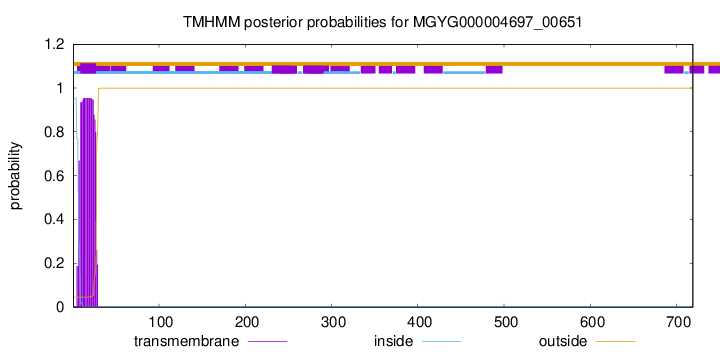
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; | |||||||||||
| CAZyme ID | MGYG000004697_00651 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13833; End: 15992 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 203 | 443 | 4.5e-86 | 0.9915611814345991 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.09e-64 | 202 | 451 | 2 | 272 | Cellulase (glycosyl hydrolase family 5). |
| sd00036 | LRR_3 | 5.07e-12 | 605 | 693 | 17 | 93 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 5.12e-12 | 605 | 693 | 63 | 139 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| COG2730 | BglC | 5.63e-12 | 179 | 421 | 33 | 330 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam13306 | LRR_5 | 3.61e-11 | 605 | 693 | 14 | 89 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAC06196.1 | 3.13e-104 | 171 | 484 | 15 | 338 |
| AGH41463.1 | 5.49e-100 | 171 | 484 | 222 | 545 |
| AAC06197.1 | 1.80e-96 | 166 | 484 | 44 | 372 |
| AGH40913.1 | 3.58e-96 | 166 | 484 | 63 | 391 |
| CBK74991.1 | 7.90e-96 | 172 | 499 | 32 | 371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 3.19e-86 | 185 | 484 | 5 | 301 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZB_A | 1.23e-80 | 183 | 479 | 2 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 3PZT_A | 2.59e-80 | 185 | 479 | 29 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 2.08e-76 | 185 | 479 | 4 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 1H11_A | 2.13e-76 | 184 | 484 | 3 | 301 | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYMEINTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION [Salipaludibacillus agaradhaerens],1H2J_A ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION [Salipaludibacillus agaradhaerens],1HF6_A ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE [Salipaludibacillus agaradhaerens],1OCQ_A COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine [Salipaludibacillus agaradhaerens],1W3K_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellobio Derived-tetrahydrooxazine [Salipaludibacillus agaradhaerens],1W3L_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellotri Derived-Tetrahydrooxazine [Salipaludibacillus agaradhaerens],4A3H_A 2',4' Dinitrophenyl-2-Deoxy-2-Fluro-B-D-Cellobioside Complex Of The Endoglucanase Cel5a From Bacillus Agaradhaerens At 1.6 A Resolution [Salipaludibacillus agaradhaerens],5A3H_A 2-Deoxy-2-Fluro-B-D-CellobiosylENZYME INTERMEDIATE COMPLEX Of The Endoglucanase Cel5a From Bacillus Agaradhearans At 1.8 Angstroms Resolution [Salipaludibacillus agaradhaerens],6A3H_A 2-Deoxy-2-Fluro-B-D-CellotriosylENZYME INTERMEDIATE COMPLEX OF THE Endoglucanase Cel5a From Bacillus Agaradhearans At 1.6 Angstrom Resolution [Salipaludibacillus agaradhaerens],7A3H_A Native Endoglucanase Cel5a Catalytic Core Domain At 0.95 Angstroms Resolution [Salipaludibacillus agaradhaerens],8A3H_A Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07983 | 3.62e-79 | 185 | 492 | 34 | 338 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 1.99e-77 | 185 | 479 | 34 | 325 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P06565 | 9.34e-77 | 189 | 500 | 34 | 335 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| Q59394 | 3.62e-75 | 185 | 485 | 33 | 330 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
| Q47096 | 9.35e-75 | 185 | 485 | 33 | 330 | Endoglucanase 5 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=celV PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
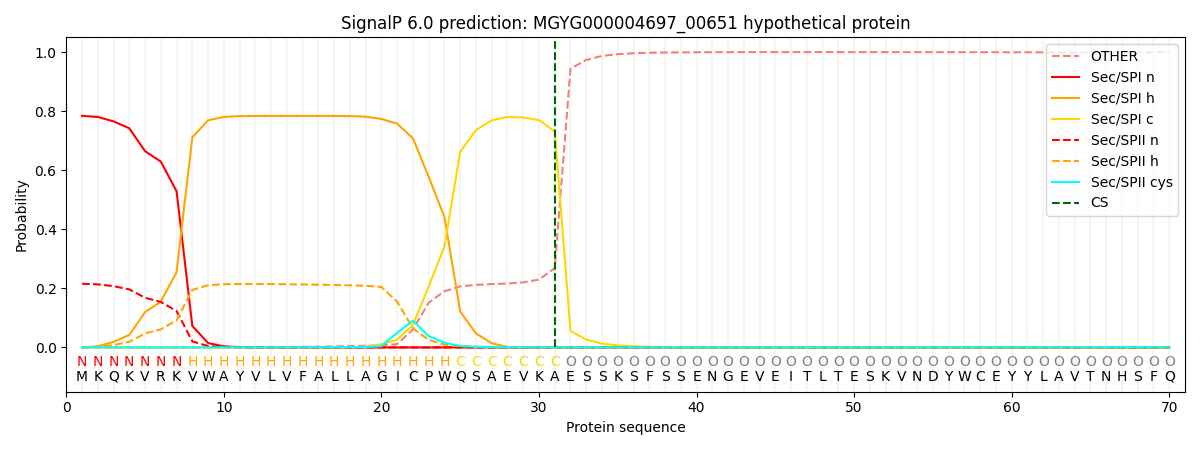
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001476 | 0.776963 | 0.220391 | 0.000592 | 0.000302 | 0.000247 |