You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004697_00957
You are here: Home > Sequence: MGYG000004697_00957
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
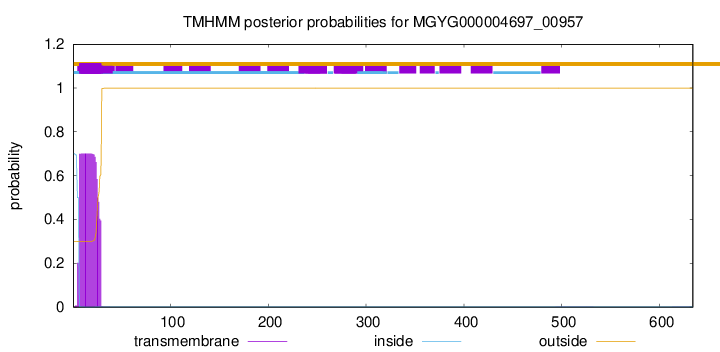
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; | |||||||||||
| CAZyme ID | MGYG000004697_00957 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29853; End: 31757 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 6.66e-23 | 393 | 622 | 45 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 9.04e-09 | 45 | 98 | 3 | 53 | Bacterial SH3 domain. |
| COG4991 | YraI | 8.12e-07 | 27 | 101 | 24 | 95 | Uncharacterized conserved protein YraI [Function unknown]. |
| COG3103 | YgiM | 1.84e-04 | 24 | 101 | 78 | 152 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| smart00287 | SH3b | 3.27e-04 | 33 | 96 | 1 | 59 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHQ61172.1 | 3.40e-126 | 2 | 634 | 3 | 676 |
| BCJ93456.1 | 7.98e-114 | 249 | 633 | 390 | 803 |
| BCN29841.1 | 2.03e-112 | 216 | 634 | 353 | 798 |
| ABX43817.1 | 2.60e-111 | 217 | 633 | 749 | 1182 |
| BBF45313.1 | 9.67e-111 | 27 | 634 | 27 | 652 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 3.01e-09 | 469 | 622 | 94 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.00e-13 | 396 | 622 | 679 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q8CPQ1 | 5.13e-10 | 396 | 622 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 2.02e-09 | 396 | 622 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 2.02e-09 | 396 | 622 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q99V41 | 9.17e-08 | 469 | 622 | 1098 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
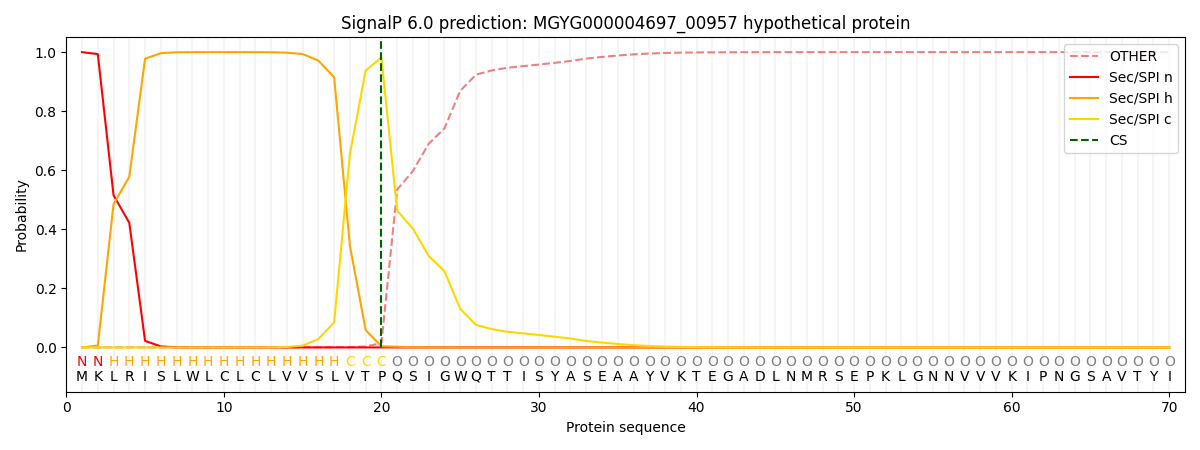
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000780 | 0.998369 | 0.000302 | 0.000175 | 0.000164 | 0.000174 |