You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004697_00967
You are here: Home > Sequence: MGYG000004697_00967
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; | |||||||||||
| CAZyme ID | MGYG000004697_00967 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42048; End: 44276 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 370 | 737 | 3.2e-74 | 0.9900990099009901 |
| CBM22 | 217 | 338 | 1.4e-27 | 0.9770992366412213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 9.05e-65 | 370 | 737 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 3.14e-62 | 456 | 735 | 14 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 5.58e-42 | 407 | 737 | 55 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 3.11e-24 | 216 | 341 | 3 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| PRK10905 | PRK10905 | 3.63e-08 | 131 | 214 | 138 | 225 | cell division protein DamX; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFU34339.1 | 3.54e-106 | 369 | 738 | 173 | 519 |
| QFJ54853.1 | 8.11e-100 | 336 | 738 | 4 | 381 |
| BCZ47872.1 | 9.16e-100 | 369 | 738 | 45 | 399 |
| CBK75021.1 | 2.01e-99 | 369 | 738 | 37 | 381 |
| AQS12857.1 | 2.31e-97 | 369 | 738 | 37 | 391 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5F_A | 6.86e-54 | 221 | 734 | 29 | 523 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 1.86e-50 | 221 | 734 | 29 | 523 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 7NL2_A | 5.65e-36 | 403 | 738 | 37 | 340 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 3W24_A | 2.54e-35 | 408 | 741 | 40 | 329 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W25_A | 1.60e-34 | 408 | 741 | 40 | 329 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W26_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23551 | 5.18e-86 | 369 | 738 | 37 | 382 | Endo-1,4-beta-xylanase A OS=Butyrivibrio fibrisolvens OX=831 GN=xynA PE=3 SV=1 |
| P51584 | 1.35e-51 | 221 | 734 | 40 | 534 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60037 | 3.13e-51 | 208 | 721 | 197 | 676 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P36917 | 3.75e-51 | 199 | 741 | 181 | 678 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| Q60042 | 3.52e-48 | 209 | 721 | 193 | 672 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
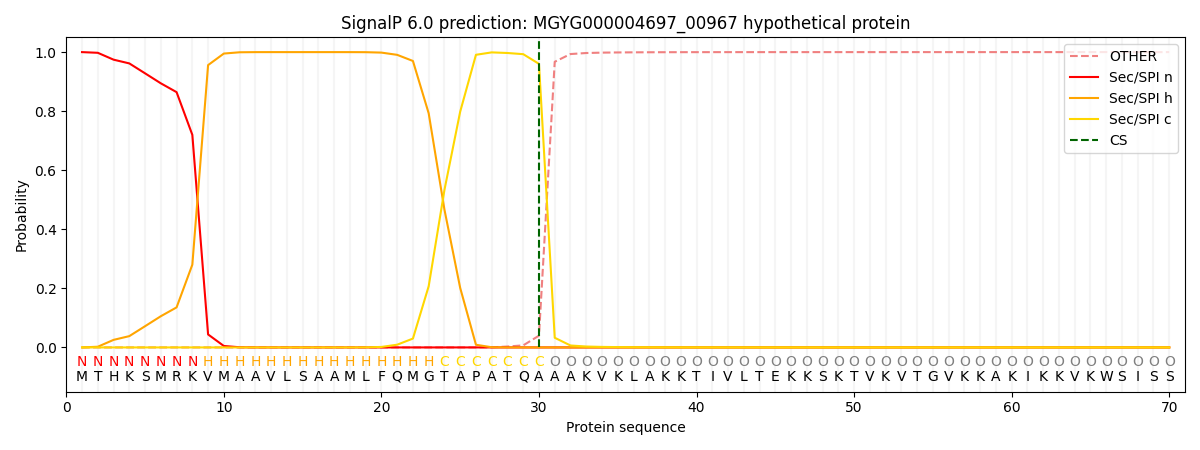
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000420 | 0.998818 | 0.000168 | 0.000214 | 0.000183 | 0.000159 |
