You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004703_00336
You are here: Home > Sequence: MGYG000004703_00336
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900556275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900556275 | |||||||||||
| CAZyme ID | MGYG000004703_00336 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57193; End: 59334 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 587 | 695 | 1.6e-21 | 0.7983870967741935 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16216 | GxGYxYP_N | 8.74e-48 | 102 | 316 | 1 | 215 | GxGYxY sequence motif in domain of unknown function N-terminal. This domain is found in bacteria, archaea and eukaryotes, and is typically between 213 and 231 amino acids in length. This domain is found in association with pfam14323. |
| pfam14323 | GxGYxYP_C | 9.98e-41 | 336 | 549 | 1 | 218 | GxGYxYP putative glycoside hydrolase C-terminal domain. This family carries a characteristic sequence motif, GxGYxYP, and is a putative glycoside hydrolase. This domain is found in association with pfam16216. Associated families are sugar-processing domains. |
| pfam00754 | F5_F8_type_C | 1.60e-13 | 600 | 694 | 19 | 113 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.08e-06 | 582 | 694 | 12 | 129 | Substituted updates: Jan 31, 2002 |
| pfam17132 | Glyco_hydro_106 | 7.59e-04 | 577 | 705 | 166 | 296 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANH82740.1 | 1.02e-203 | 54 | 712 | 24 | 680 |
| QJE98862.1 | 3.02e-162 | 66 | 711 | 41 | 698 |
| BCJ99038.1 | 5.08e-82 | 97 | 574 | 195 | 662 |
| QUT77716.1 | 4.01e-30 | 97 | 409 | 47 | 372 |
| BCI61582.1 | 7.46e-19 | 591 | 711 | 334 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3SGG_A | 4.19e-29 | 94 | 461 | 57 | 417 | Crystalstructure of a putative hydrolase (BT_2193) from Bacteroides thetaiotaomicron VPI-5482 at 1.25 A resolution [Bacteroides thetaiotaomicron] |
| 2V5D_A | 1.04e-06 | 574 | 699 | 587 | 719 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
| 2J7M_A | 3.81e-06 | 584 | 699 | 11 | 131 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 3.89e-06 | 584 | 699 | 12 | 132 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 1.59e-06 | 574 | 712 | 617 | 768 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 1.59e-06 | 574 | 712 | 617 | 768 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
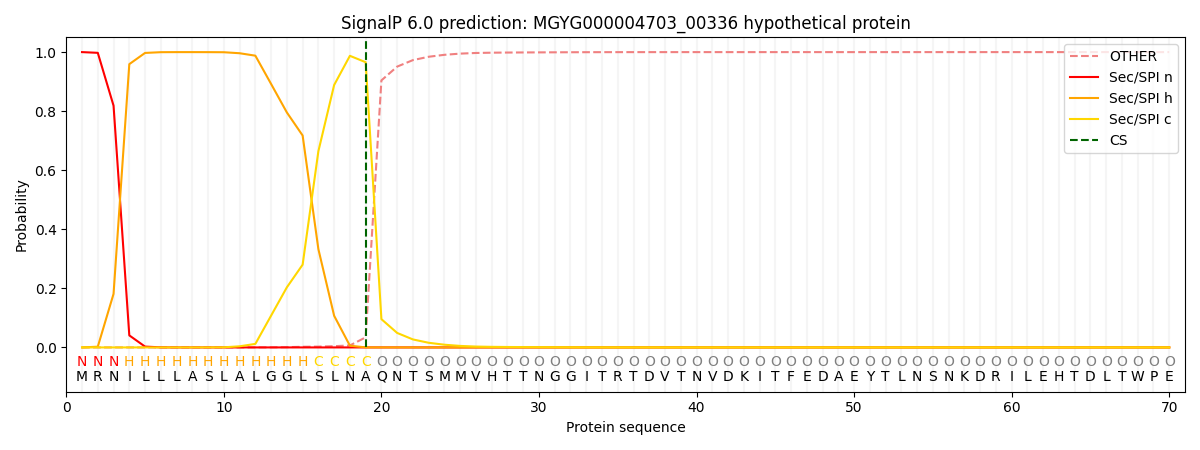
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000356 | 0.998959 | 0.000214 | 0.000161 | 0.000155 | 0.000144 |
