You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004703_00985
You are here: Home > Sequence: MGYG000004703_00985
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900556275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900556275 | |||||||||||
| CAZyme ID | MGYG000004703_00985 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 272; End: 3211 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 545 | 933 | 6.6e-38 | 0.6587301587301587 |
| CBM66 | 72 | 227 | 4.4e-23 | 0.9741935483870968 |
| CBM66 | 246 | 399 | 8.4e-19 | 0.9612903225806452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 6.00e-18 | 548 | 930 | 48 | 498 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 2.38e-16 | 745 | 924 | 3 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 1.12e-11 | 745 | 924 | 6 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06439 | DUF1080 | 6.29e-11 | 284 | 402 | 56 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| pfam06439 | DUF1080 | 2.50e-09 | 78 | 230 | 21 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP87692.1 | 1.69e-185 | 202 | 928 | 43 | 795 |
| CBW20971.1 | 2.23e-185 | 202 | 928 | 52 | 804 |
| BAD47119.1 | 3.03e-185 | 202 | 928 | 62 | 814 |
| CUA16970.1 | 4.73e-185 | 202 | 928 | 43 | 795 |
| QCQ40063.1 | 6.02e-185 | 202 | 928 | 62 | 814 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8VZR2 | 2.10e-28 | 449 | 930 | 139 | 652 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q9SG80 | 4.48e-26 | 445 | 930 | 134 | 655 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| U6A629 | 7.43e-16 | 420 | 692 | 73 | 391 | Alpha-L-arabinofuranosidase A OS=Penicillium canescens OX=5083 GN=abfA PE=1 SV=1 |
| Q0CTV2 | 5.12e-15 | 420 | 914 | 78 | 607 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfA PE=3 SV=1 |
| Q96X54 | 7.37e-13 | 420 | 914 | 78 | 607 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus awamori OX=105351 GN=abfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
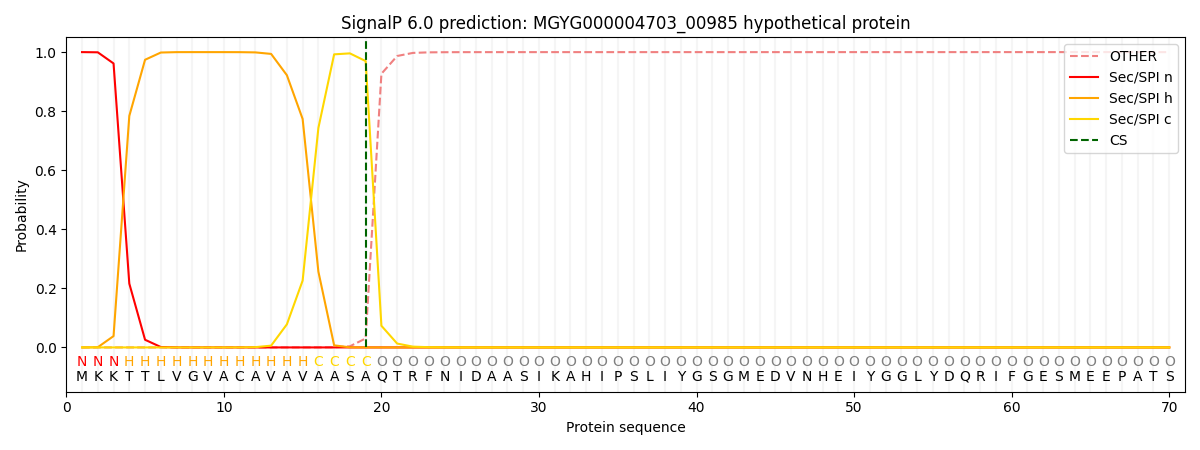
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000319 | 0.998851 | 0.000237 | 0.000178 | 0.000199 | 0.000176 |
