You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004707_01087
You are here: Home > Sequence: MGYG000004707_01087
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
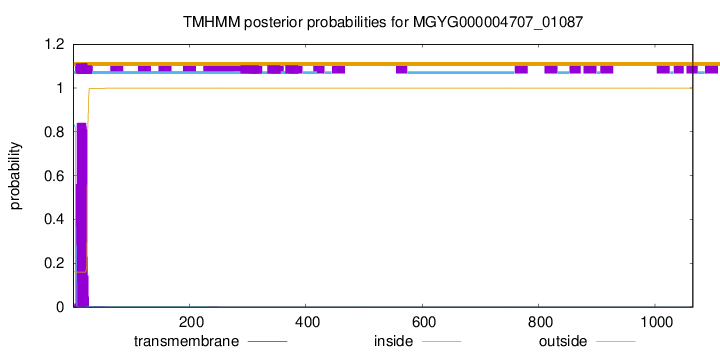
TMHMM annotations
Basic Information help
| Species | Monoglobus sp900554105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus sp900554105 | |||||||||||
| CAZyme ID | MGYG000004707_01087 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17431; End: 20628 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 94 | 310 | 1.6e-67 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PHA03247 | PHA03247 | 3.66e-07 | 882 | 975 | 2690 | 2785 | large tegument protein UL36; Provisional |
| PHA03247 | PHA03247 | 1.73e-06 | 892 | 996 | 2704 | 2813 | large tegument protein UL36; Provisional |
| NF033761 | gliding_GltJ | 3.45e-06 | 894 | 979 | 394 | 482 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| PRK12270 | kgd | 3.63e-06 | 903 | 1001 | 37 | 133 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| NF033761 | gliding_GltJ | 6.77e-06 | 887 | 993 | 406 | 512 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19402.1 | 1.22e-151 | 1 | 522 | 1 | 494 |
| AUO19400.1 | 1.74e-147 | 1 | 522 | 1 | 500 |
| AUO19476.1 | 1.69e-119 | 39 | 531 | 33 | 485 |
| AUO19050.1 | 1.11e-89 | 39 | 531 | 47 | 533 |
| AXP81340.1 | 2.10e-89 | 39 | 526 | 40 | 506 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B0XMA2 | 1.03e-53 | 40 | 526 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.40e-53 | 40 | 526 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| A1DPF0 | 2.58e-53 | 40 | 526 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 5.43e-52 | 40 | 529 | 20 | 417 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 3.95e-50 | 40 | 529 | 20 | 417 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
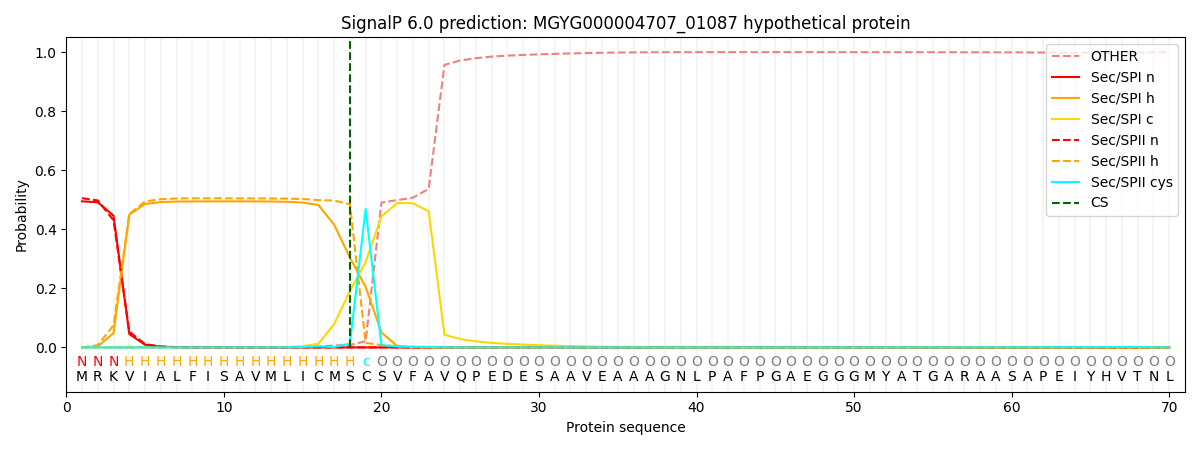
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000557 | 0.485589 | 0.513156 | 0.000274 | 0.000226 | 0.000188 |