You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004708_00053
You are here: Home > Sequence: MGYG000004708_00053
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004708_00053 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29845; End: 32496 Strand: + | |||||||||||
Full Sequence Download help
| MSVISKCILF LSCLCVSLQS QAQLWYDMKP EMRPGTRWWW HGSAVTKEGI DHELDEFNKK | 60 |
| GIVSVEITPI HGVIGNEKNE LDFLSPQWMD VLDHTVRSAV RRGMRADMTT GTGWPFGGPW | 120 |
| VRPEDGACAA EFVTDTINRK SDIKGLWAEL LDCRLLRLRQ RNNAKLSYVV AHKLCKDGSL | 180 |
| GKAKDITHLV IEGKPLRCGN KDSVLVIALY IVRTGQQVER AAPNGEGLVI DHFSGKAVTD | 240 |
| YLEVFDKSFD TGHTAVPHSF FCDSYEVKRA NWTPELLDTF YKIKGYRLEE HLPELLGLYG | 300 |
| DNSVLCDYRE TLGNMLLDNF SKKWASWARR KGASVRYQAH GSPGNLIDLY AAADIPETEG | 360 |
| FGLTDFNIKG LRRDDGFTRR NLSDVSMYKY ASSAANVTSK PYTSAETFTW LTEHFRTSLS | 420 |
| QMKPELDLMF TCGINRIMFH GSSYSPEYEA WPGRKFYASV DVSPTNSIWR DLCFMTKYIE | 480 |
| RCLNILQSGR QDNDFIVYLP IRNMWMKREK PGTDGLLMPF TISSARSLAP EFVKTVLDID | 540 |
| SLGFDFDYIS DRLLMQVTFR NGRLVTASGS EYKGLVIPEG SILTAEMRGH IDNLRSMGAP | 600 |
| VVFGVDKKQL ASFATAEDIR LVSGVRAIRK RLENGYGYFI ANLSGSDVDE WMSPAVSFED | 660 |
| AVWLDPMTGE DFACEKDGRG IHIVLRSGES ILLRTFDSRL INRNRKSRPA GKVCGEIDLT | 720 |
| DNGWQLMFVD EVPCVRDTFN LASLSSWEKL NDDSVRVTMG TGVYTTVFQL TGKQPEQVRE | 780 |
| IDLGDVRESA RVYINGHFIG CAWAAPFILR IDGGILKTGN NELRIEVTNL PANRIAEMDR | 840 |
| KGITWRKFKE INFVDLHYNR NSRYSRWLPV ASGLNSRVRL LLE | 883 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 22 | 617 | 6e-152 | 0.6225728155339806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 3.24e-36 | 214 | 670 | 385 | 865 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85006.1 | 6.64e-293 | 13 | 876 | 10 | 834 |
| QJR98068.1 | 1.10e-289 | 31 | 880 | 6 | 859 |
| QVJ80546.1 | 1.03e-287 | 10 | 881 | 4 | 803 |
| ADE82941.1 | 8.31e-287 | 10 | 881 | 4 | 803 |
| AGB29019.1 | 3.99e-258 | 20 | 880 | 19 | 854 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 2.67e-33 | 222 | 839 | 412 | 1065 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 6.13e-33 | 222 | 839 | 412 | 1065 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6Q2F_A | 5.81e-29 | 221 | 845 | 455 | 1115 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 3.60e-12 | 669 | 842 | 926 | 1109 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
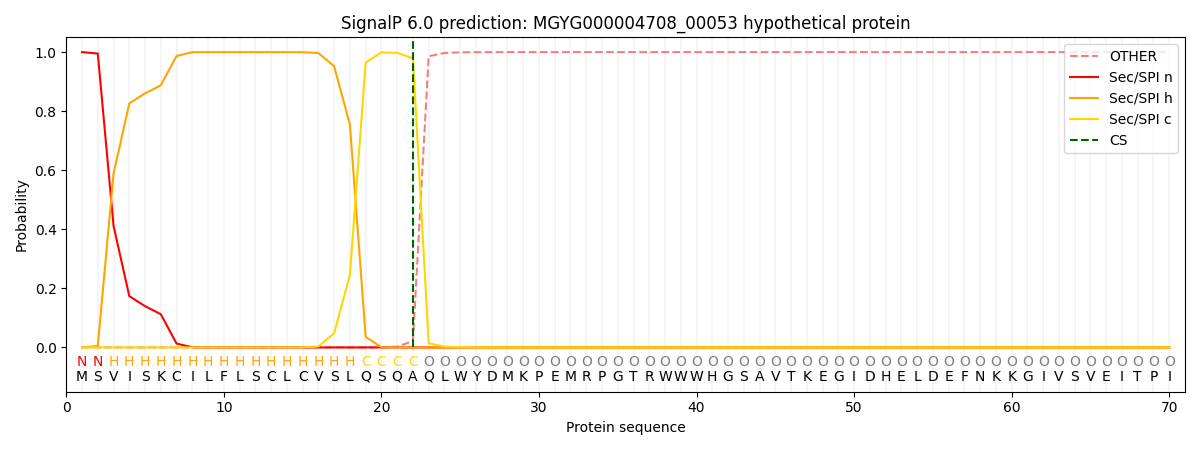
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000366 | 0.998878 | 0.000271 | 0.000163 | 0.000149 | 0.000140 |
