You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004721_00949
You are here: Home > Sequence: MGYG000004721_00949
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004721_00949 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | Pectinesterase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 728; End: 2074 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 42 | 315 | 5.5e-35 | 0.8680555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.67e-15 | 33 | 316 | 81 | 378 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 1.12e-08 | 101 | 371 | 55 | 291 | putative pectinesterase |
| PLN02488 | PLN02488 | 2.80e-08 | 28 | 368 | 192 | 495 | probable pectinesterase/pectinesterase inhibitor |
| PLN02773 | PLN02773 | 1.02e-07 | 151 | 313 | 107 | 268 | pectinesterase |
| PLN02497 | PLN02497 | 8.88e-07 | 173 | 313 | 149 | 292 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65238.1 | 7.14e-166 | 28 | 443 | 661 | 1072 |
| QCD41145.1 | 4.87e-141 | 28 | 445 | 1033 | 1437 |
| QCP72097.1 | 6.37e-130 | 30 | 447 | 21 | 428 |
| QCD38407.1 | 6.37e-130 | 30 | 447 | 21 | 428 |
| AEF12641.1 | 5.43e-107 | 24 | 441 | 1033 | 1442 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LUL8 | 2.42e-07 | 86 | 381 | 691 | 967 | Putative pectinesterase/pectinesterase inhibitor 26 OS=Arabidopsis thaliana OX=3702 GN=PME26 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
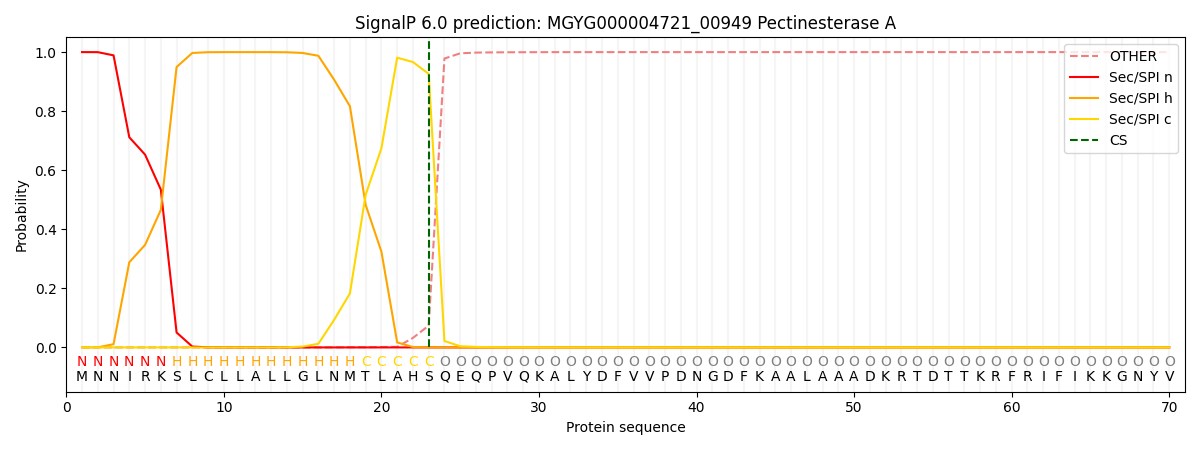
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000355 | 0.998890 | 0.000253 | 0.000175 | 0.000157 | 0.000152 |
