You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004721_00950
You are here: Home > Sequence: MGYG000004721_00950
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004721_00950 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2152; End: 6669 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 2.73e-14 | 1007 | 1300 | 53 | 355 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 5.96e-13 | 1101 | 1350 | 41 | 293 | pectinesterase |
| PLN02170 | PLN02170 | 6.17e-12 | 1046 | 1379 | 226 | 519 | probable pectinesterase/pectinesterase inhibitor |
| PLN02432 | PLN02432 | 7.43e-12 | 1164 | 1331 | 101 | 265 | putative pectinesterase |
| pfam01095 | Pectinesterase | 8.03e-11 | 1046 | 1333 | 1 | 273 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD41145.1 | 0.0 | 9 | 1502 | 10 | 1492 |
| QNT65238.1 | 0.0 | 393 | 1502 | 13 | 1130 |
| QUT74167.1 | 0.0 | 26 | 1501 | 32 | 1431 |
| QCD38476.1 | 0.0 | 22 | 1502 | 21 | 1473 |
| QCP72166.1 | 0.0 | 22 | 1502 | 21 | 1473 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 4.34e-10 | 1040 | 1307 | 26 | 308 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SMY6 | 1.14e-06 | 1053 | 1299 | 305 | 536 | Putative pectinesterase/pectinesterase inhibitor 45 OS=Arabidopsis thaliana OX=3702 GN=PME45 PE=2 SV=1 |
| Q7Y201 | 3.41e-06 | 1004 | 1345 | 283 | 585 | Probable pectinesterase/pectinesterase inhibitor 13 OS=Arabidopsis thaliana OX=3702 GN=PME13 PE=2 SV=2 |
| Q4PSN0 | 5.71e-06 | 1186 | 1302 | 153 | 272 | Probable pectinesterase 29 OS=Arabidopsis thaliana OX=3702 GN=PME29 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
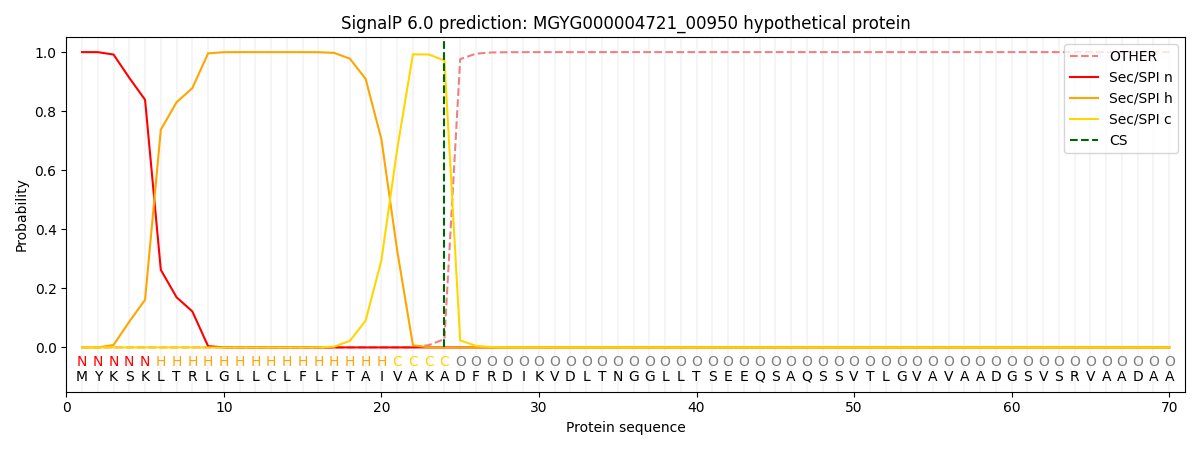
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000361 | 0.998703 | 0.000354 | 0.000170 | 0.000183 | 0.000171 |
