You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004731_00826
You are here: Home > Sequence: MGYG000004731_00826
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bifidobacterium italicum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium italicum | |||||||||||
| CAZyme ID | MGYG000004731_00826 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1460; End: 2356 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 94 | 291 | 1.5e-35 | 0.9841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01083 | Cutinase | 9.30e-19 | 93 | 292 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATU20358.1 | 7.84e-151 | 1 | 298 | 1 | 299 |
| ASW23299.1 | 1.17e-81 | 20 | 296 | 11 | 297 |
| AIZ15958.1 | 4.65e-80 | 75 | 296 | 2 | 228 |
| ATO40344.1 | 9.26e-80 | 75 | 296 | 23 | 249 |
| AFI62706.1 | 4.07e-65 | 75 | 296 | 79 | 305 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2AXE_A | 3.46e-09 | 90 | 297 | 1 | 207 | IodinatedComplex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
| 1BS9_A | 4.70e-09 | 90 | 297 | 1 | 207 | AcetylxylanEsterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
| 1G66_A | 1.60e-08 | 91 | 297 | 2 | 207 | ACETYLXYLANESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
| 1QOZ_A | 9.90e-08 | 126 | 296 | 47 | 206 | ChainA, ACETYL XYLAN ESTERASE [Trichoderma reesei],1QOZ_B Chain B, ACETYL XYLAN ESTERASE [Trichoderma reesei] |
| 4PSC_A | 1.52e-07 | 96 | 218 | 81 | 196 | Structureof cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WP43 | 5.38e-10 | 85 | 292 | 26 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut7 PE=1 SV=1 |
| P63880 | 5.38e-10 | 85 | 292 | 26 | 213 | Carboxylesterase Culp1 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2006C PE=3 SV=1 |
| P9WP42 | 5.38e-10 | 85 | 292 | 26 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2037 PE=3 SV=1 |
| A6WFI5 | 1.14e-09 | 95 | 295 | 37 | 221 | Cutinase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=cut PE=1 SV=1 |
| O59893 | 3.49e-08 | 90 | 297 | 28 | 234 | Acetylxylan esterase 2 OS=Talaromyces purpureogenus OX=1266744 GN=axe-2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
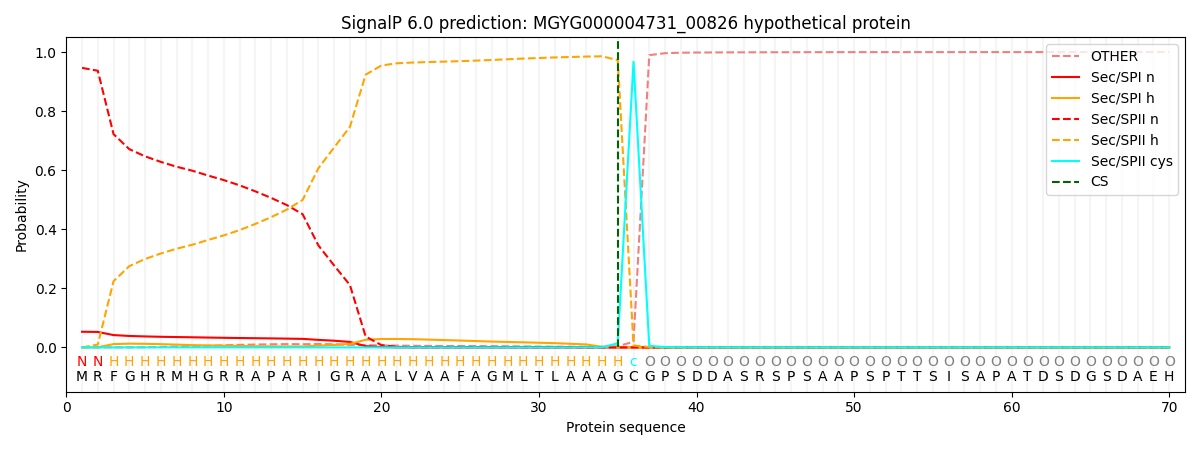
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000089 | 0.020262 | 0.979625 | 0.000023 | 0.000015 | 0.000003 |
