You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004739_00354
You are here: Home > Sequence: MGYG000004739_00354
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Actinomyces sp000220835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; Actinomyces sp000220835 | |||||||||||
| CAZyme ID | MGYG000004739_00354 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22377; End: 23324 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 65 | 192 | 3e-28 | 0.8222222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13399 | Slt35-like | 3.81e-34 | 66 | 165 | 3 | 97 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| cd00254 | LT-like | 1.17e-26 | 68 | 189 | 1 | 110 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| pfam00877 | NLPC_P60 | 1.88e-18 | 216 | 299 | 1 | 90 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 5.21e-18 | 205 | 300 | 76 | 180 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| cd13401 | Slt70-like | 2.17e-17 | 49 | 193 | 3 | 150 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEI18427.1 | 3.08e-126 | 41 | 315 | 35 | 308 |
| VEG28534.1 | 2.18e-113 | 48 | 314 | 271 | 534 |
| CED91304.1 | 4.04e-90 | 20 | 312 | 12 | 319 |
| VEG29992.1 | 9.42e-85 | 15 | 312 | 1 | 314 |
| AYD89184.1 | 1.95e-67 | 10 | 303 | 5 | 307 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XIV_A | 9.27e-16 | 205 | 312 | 81 | 204 | Structureof Rv1477, hypothetical invasion protein of Mycobacterium tuberculosis [Mycobacterium tuberculosis H37Rv] |
| 3PBC_A | 1.02e-15 | 205 | 312 | 86 | 209 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
| 3NE0_A | 1.02e-15 | 205 | 312 | 86 | 209 | Structureand functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation [Mycobacterium tuberculosis H37Rv] |
| 3S0Q_A | 1.79e-14 | 205 | 312 | 87 | 210 | ChainA, INVASION PROTEIN [Mycobacterium tuberculosis] |
| 4Q4T_A | 4.98e-14 | 205 | 312 | 344 | 467 | Structureof the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WHU3 | 6.85e-16 | 198 | 311 | 268 | 380 | Probable endopeptidase Rv2190c OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv2190c PE=3 SV=1 |
| P9WHU2 | 6.85e-16 | 198 | 311 | 268 | 380 | Probable endopeptidase MT2245 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2245 PE=3 SV=1 |
| P67474 | 6.85e-16 | 198 | 311 | 268 | 380 | Probable endopeptidase Mb2213c OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2213C PE=3 SV=1 |
| O31976 | 1.67e-15 | 37 | 191 | 1407 | 1543 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
| O64046 | 1.67e-15 | 37 | 191 | 1407 | 1543 | Probable tape measure protein OS=Bacillus phage SPbeta OX=66797 GN=yomI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
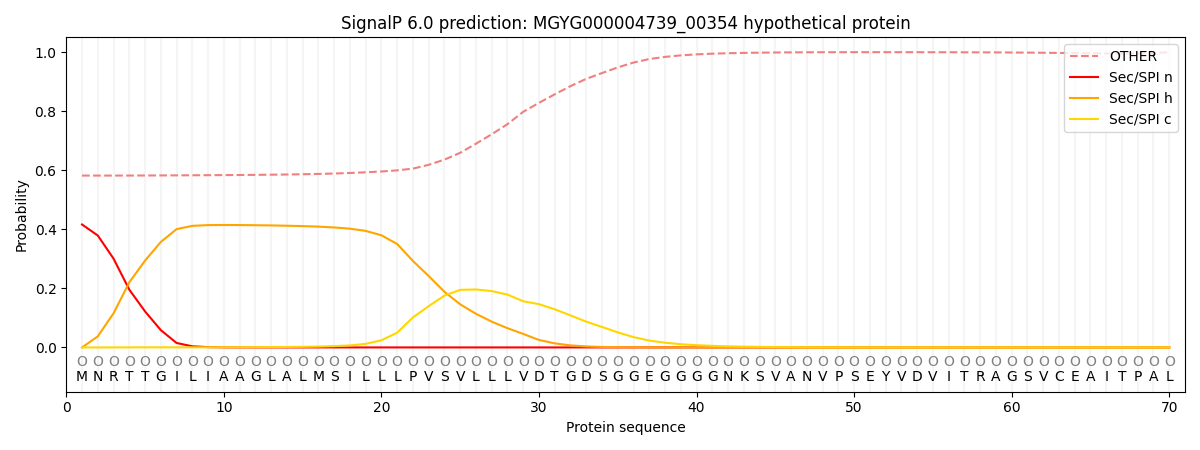
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.599693 | 0.394246 | 0.001128 | 0.001245 | 0.000754 | 0.002935 |

