You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004739_00374
You are here: Home > Sequence: MGYG000004739_00374
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Actinomyces sp000220835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; Actinomyces sp000220835 | |||||||||||
| CAZyme ID | MGYG000004739_00374 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49876; End: 52173 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00701 | PGRP | 4.88e-38 | 292 | 432 | 1 | 132 | Animal peptidoglycan recognition proteins homologous to Bacteriophage T3 lysozyme. The bacteriophage molecule, but not its moth homologue, has been shown to have N-acetylmuramoyl-L-alanine amidase activity. One member of this family, Tag7, is a cytokine. |
| cd06583 | PGRP | 1.12e-27 | 317 | 472 | 3 | 125 | Peptidoglycan recognition proteins (PGRPs) are pattern recognition receptors that bind, and in certain cases, hydrolyze peptidoglycans (PGNs) of bacterial cell walls. PGRPs have been divided into three classes: short PGRPs (PGRP-S), that are small (20 kDa) extracellular proteins; intermediate PGRPs (PGRP-I) that are 40-45 kDa and are predicted to be transmembrane proteins; and long PGRPs (PGRP-L), up to 90 kDa, which may be either intracellular or transmembrane. Several structures of PGRPs are known in insects and mammals, some bound with substrates like Muramyl Tripeptide (MTP) or Tracheal Cytotoxin (TCT). The substrate binding site is conserved in PGRP-LCx, PGRP-LE, and PGRP-Ialpha proteins. This family includes Zn-dependent N-Acetylmuramoyl-L-alanine Amidase, EC:3.5.1.28. This enzyme cleaves the amide bond between N-acetylmuramoyl and L-amino acids, preferentially D-lactyl-L-Ala, in bacterial cell walls. The structure for the bacteriophage T7 lysozyme shows that two of the conserved histidines and a cysteine are zinc binding residues. Site-directed mutagenesis of T7 lysozyme indicates that two conserved residues, a Tyr and a Lys, are important for amidase activity. |
| NF033930 | pneumo_PspA | 8.79e-22 | 495 | 563 | 445 | 512 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| NF033838 | PspC_subgroup_1 | 1.99e-21 | 495 | 563 | 488 | 555 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| pfam01510 | Amidase_2 | 4.20e-21 | 317 | 472 | 3 | 121 | N-acetylmuramoyl-L-alanine amidase. This family includes zinc amidases that have N-acetylmuramoyl-L-alanine amidase activity EC:3.5.1.28. This enzyme domain cleaves the amide bond between N-acetylmuramoyl and L-amino acids in bacterial cell walls (preferentially: D-lactyl-L-Ala). The structure is known for the bacteriophage T7 structure and shows that two of the conserved histidines are zinc binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEI16134.1 | 8.55e-222 | 37 | 765 | 90 | 984 |
| QPL05816.1 | 1.72e-199 | 3 | 760 | 22 | 1001 |
| VEG75872.1 | 2.44e-197 | 9 | 765 | 7 | 927 |
| AMD86541.1 | 5.48e-193 | 1 | 765 | 1 | 975 |
| AVM62882.1 | 3.47e-78 | 565 | 764 | 189 | 405 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Z8I_A | 1.03e-15 | 294 | 475 | 66 | 216 | Crystalstructure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 [Branchiostoma belcheri tsingtauense] |
| 4ZXM_A | 1.47e-15 | 294 | 475 | 95 | 245 | Crystalstructure of PGRP domain from Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 [Branchiostoma belcheri tsingtauense] |
| 2F2L_X | 2.77e-15 | 294 | 414 | 3 | 115 | Crystalstructure of tracheal cytotoxin (TCT) bound to the ectodomain complex of peptidoglycan recognition proteins LCa (PGRP-LCa) and LCx (PGRP-LCx) [Drosophila melanogaster] |
| 1YCK_A | 2.10e-12 | 294 | 475 | 12 | 163 | Crystalstructure of human peptidoglycan recognition protein (PGRP-S) [Homo sapiens] |
| 2R2K_A | 1.22e-11 | 288 | 472 | 1 | 157 | Crystalstructure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution [Camelus dromedarius],2R2K_B Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution [Camelus dromedarius],2R2K_C Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution [Camelus dromedarius],2R2K_D Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution [Camelus dromedarius],2R90_A Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution [Camelus dromedarius],2R90_B Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution [Camelus dromedarius],2R90_C Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution [Camelus dromedarius],2R90_D Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution [Camelus dromedarius],2Z9N_A Crystal structure of cameline peptidoglycan recognition protein at 3.2 A resolution [Camelus dromedarius],2Z9N_B Crystal structure of cameline peptidoglycan recognition protein at 3.2 A resolution [Camelus dromedarius],2Z9N_C Crystal structure of cameline peptidoglycan recognition protein at 3.2 A resolution [Camelus dromedarius],2Z9N_D Crystal structure of cameline peptidoglycan recognition protein at 3.2 A resolution [Camelus dromedarius],3C2X_A Crystal structure of peptidoglycan recognition protein at 1.8A resolution [Camelus dromedarius],3C2X_B Crystal structure of peptidoglycan recognition protein at 1.8A resolution [Camelus dromedarius],3C2X_C Crystal structure of peptidoglycan recognition protein at 1.8A resolution [Camelus dromedarius],3C2X_D Crystal structure of peptidoglycan recognition protein at 1.8A resolution [Camelus dromedarius],3CG9_A Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution [Camelus dromedarius],3CG9_B Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution [Camelus dromedarius],3CG9_C Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution [Camelus dromedarius],3CG9_D Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution [Camelus dromedarius],3COR_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution [Camelus dromedarius],3COR_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution [Camelus dromedarius],3COR_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution [Camelus dromedarius],3COR_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution [Camelus dromedarius],3CXA_A Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution [Camelus dromedarius],3CXA_B Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution [Camelus dromedarius],3CXA_C Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution [Camelus dromedarius],3CXA_D Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution [Camelus dromedarius],3NG4_A Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution [Camelus dromedarius],3NG4_B Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution [Camelus dromedarius],3NG4_C Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution [Camelus dromedarius],3NG4_D Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution [Camelus dromedarius],3NNO_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution [Camelus dromedarius],3NNO_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution [Camelus dromedarius],3NNO_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution [Camelus dromedarius],3NNO_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with Alpha-Rhamnose at 2.9 A resolution [Camelus dromedarius],3NW3_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution [Camelus dromedarius],3NW3_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution [Camelus dromedarius],3NW3_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution [Camelus dromedarius],3NW3_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution [Camelus dromedarius],3O4K_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution [Camelus dromedarius],3O4K_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution [Camelus dromedarius],3O4K_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution [Camelus dromedarius],3O4K_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution [Camelus dromedarius],3OGX_A Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution [Camelus dromedarius],3OGX_B Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution [Camelus dromedarius],3OGX_C Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution [Camelus dromedarius],3OGX_D Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution [Camelus dromedarius],3QJ1_A Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution [Camelus dromedarius],3QJ1_B Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution [Camelus dromedarius],3QJ1_C Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution [Camelus dromedarius],3QJ1_D Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution [Camelus dromedarius],3QS0_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution [Camelus dromedarius],3QS0_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution [Camelus dromedarius],3QS0_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution [Camelus dromedarius],3QS0_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution [Camelus dromedarius],3QV4_A Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution [Camelus dromedarius],3QV4_B Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution [Camelus dromedarius],3QV4_C Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution [Camelus dromedarius],3QV4_D Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution [Camelus dromedarius],3RT4_A Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein [Camelus dromedarius],3RT4_B Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein [Camelus dromedarius],3RT4_C Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein [Camelus dromedarius],3RT4_D Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein [Camelus dromedarius],3T2V_A Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution [Camelus dromedarius],3T2V_B Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution [Camelus dromedarius],3T2V_C Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution [Camelus dromedarius],3T2V_D Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution [Camelus dromedarius],3T39_A Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution [Camelus dromedarius],3T39_B Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution [Camelus dromedarius],3T39_C Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution [Camelus dromedarius],3T39_D Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution [Camelus dromedarius],3TRU_A Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution [Camelus dromedarius],3TRU_B Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution [Camelus dromedarius],3TRU_C Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution [Camelus dromedarius],3TRU_D Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution [Camelus dromedarius],3UIL_A Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution [Camelus dromedarius],3UIL_B Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution [Camelus dromedarius],3UIL_C Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution [Camelus dromedarius],3UIL_D Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution [Camelus dromedarius],3UML_A Crystal structure of PGRP-S complexed with chorismate at 2.15 A resolution [Camelus dromedarius],3UML_B Crystal structure of PGRP-S complexed with chorismate at 2.15 A resolution [Camelus dromedarius],3UML_C Crystal structure of PGRP-S complexed with chorismate at 2.15 A resolution [Camelus dromedarius],3UML_D Crystal structure of PGRP-S complexed with chorismate at 2.15 A resolution [Camelus dromedarius],3UMQ_A Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution [Camelus dromedarius],3UMQ_B Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution [Camelus dromedarius],3UMQ_C Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution [Camelus dromedarius],3UMQ_D Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution [Camelus dromedarius],3USX_A Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution [Camelus dromedarius],3USX_B Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution [Camelus dromedarius],3USX_C Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution [Camelus dromedarius],3USX_D Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution [Camelus dromedarius],4FNN_A Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION [Camelus dromedarius],4FNN_B Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION [Camelus dromedarius],4FNN_C Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION [Camelus dromedarius],4FNN_D Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION [Camelus dromedarius],4GF9_A Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid [Camelus dromedarius],4GF9_B Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid [Camelus dromedarius],4GF9_C Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid [Camelus dromedarius],4GF9_D Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid [Camelus dromedarius],4OPP_A Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution [Camelus dromedarius],4OPP_B Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution [Camelus dromedarius],4OPP_C Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution [Camelus dromedarius],4OPP_D Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution [Camelus dromedarius],4ORV_A Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution [Camelus dromedarius],4ORV_B Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution [Camelus dromedarius],4ORV_C Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution [Camelus dromedarius],4ORV_D Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution [Camelus dromedarius],4OUG_A Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution [Camelus dromedarius],4OUG_B Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution [Camelus dromedarius],4OUG_C Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution [Camelus dromedarius],4OUG_D Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution [Camelus dromedarius],4Q8S_A Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution [Camelus dromedarius],4Q8S_B Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution [Camelus dromedarius],4Q8S_C Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution [Camelus dromedarius],4Q8S_D Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution [Camelus dromedarius],4Q9E_A Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution [Camelus dromedarius],4Q9E_B Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution [Camelus dromedarius],4Q9E_C Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution [Camelus dromedarius],4Q9E_D Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution [Camelus dromedarius],5E0A_A Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A [Camelus dromedarius],5E0A_B Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A [Camelus dromedarius],5E0A_C Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A [Camelus dromedarius],5E0A_D Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A [Camelus dromedarius],5E0B_A Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution [Camelus dromedarius],5E0B_B Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution [Camelus dromedarius],5E0B_C Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution [Camelus dromedarius],5E0B_D Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution [Camelus dromedarius],5XGY_A Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution [Camelus dromedarius],5XGY_B Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution [Camelus dromedarius],5XGY_C Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution [Camelus dromedarius],5XGY_D Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution [Camelus dromedarius],6J3W_A Crystal structure of Peptidoglycan recognition protein from camel with two specific tartrate binding sites at 2.07 A resolution. [Camelus dromedarius],6J3W_B Crystal structure of Peptidoglycan recognition protein from camel with two specific tartrate binding sites at 2.07 A resolution. [Camelus dromedarius],6J3W_C Crystal structure of Peptidoglycan recognition protein from camel with two specific tartrate binding sites at 2.07 A resolution. [Camelus dromedarius],6J3W_D Crystal structure of Peptidoglycan recognition protein from camel with two specific tartrate binding sites at 2.07 A resolution. [Camelus dromedarius],6JVE_A Crystal Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with tartaric acid and hexanoic acid at 2.28 A resolution [Camelus dromedarius],6JVE_B Crystal Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with tartaric acid and hexanoic acid at 2.28 A resolution [Camelus dromedarius],6JVE_C Crystal Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with tartaric acid and hexanoic acid at 2.28 A resolution [Camelus dromedarius],6JVE_D Crystal Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with tartaric acid and hexanoic acid at 2.28 A resolution [Camelus dromedarius],6JX8_A Crystal Structure of the complex of peptidoglycan recognition protein, PGRP-S from Camelus dromedarius at 2.67 A resolution. [Camelus dromedarius],6JX8_B Crystal Structure of the complex of peptidoglycan recognition protein, PGRP-S from Camelus dromedarius at 2.67 A resolution. [Camelus dromedarius],6JX8_C Crystal Structure of the complex of peptidoglycan recognition protein, PGRP-S from Camelus dromedarius at 2.67 A resolution. [Camelus dromedarius],6JX8_D Crystal Structure of the complex of peptidoglycan recognition protein, PGRP-S from Camelus dromedarius at 2.67 A resolution. [Camelus dromedarius],7DY5_A Chain A, Peptidoglycan recognition protein 1 [Camelus dromedarius],7DY5_B Chain B, Peptidoglycan recognition protein 1 [Camelus dromedarius],7DY5_C Chain C, Peptidoglycan recognition protein 1 [Camelus dromedarius],7DY5_D Chain D, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFW_A Chain A, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFW_B Chain B, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFW_C Chain C, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFW_D Chain D, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFX_A Chain A, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFX_B Chain B, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFX_C Chain C, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFX_D Chain D, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFY_A Chain A, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFY_B Chain B, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFY_C Chain C, Peptidoglycan recognition protein 1 [Camelus dromedarius],7XFY_D Chain D, Peptidoglycan recognition protein 1 [Camelus dromedarius] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q70PU1 | 3.03e-18 | 292 | 471 | 21 | 169 | Peptidoglycan-recognition protein SC2 OS=Drosophila simulans OX=7240 GN=PGRP-SC2 PE=3 SV=1 |
| Q9V4X2 | 1.41e-17 | 292 | 471 | 21 | 169 | Peptidoglycan-recognition protein SC2 OS=Drosophila melanogaster OX=7227 GN=PGRP-SC2 PE=2 SV=1 |
| O51481 | 4.78e-16 | 634 | 743 | 77 | 182 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
| Q8SPP7 | 5.45e-13 | 294 | 480 | 27 | 183 | Peptidoglycan recognition protein 1 OS=Bos taurus OX=9913 GN=PGLYRP1 PE=1 SV=1 |
| B5T255 | 2.51e-12 | 294 | 480 | 27 | 183 | Peptidoglycan recognition protein 1 OS=Bos indicus OX=9915 GN=PGLYRP1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
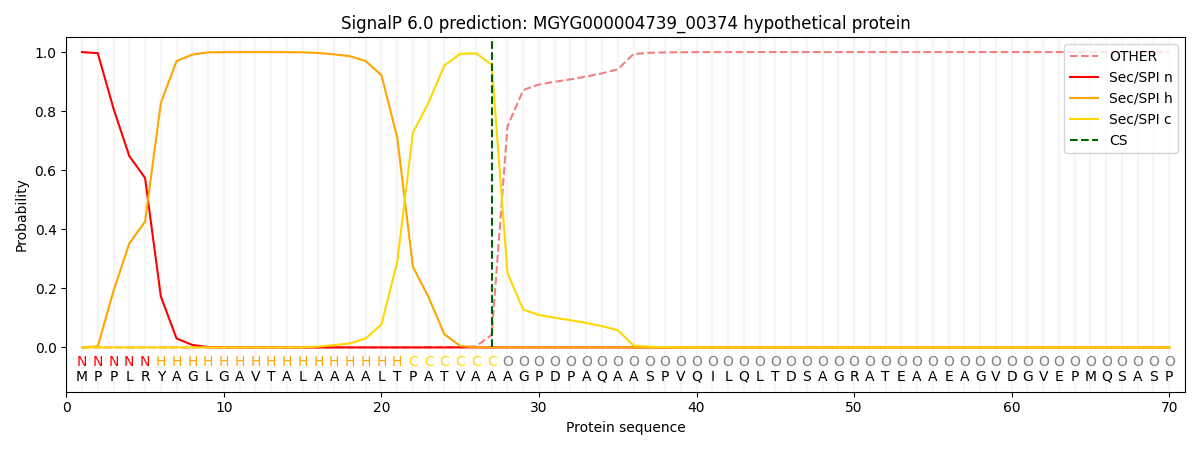
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000536 | 0.998658 | 0.000206 | 0.000230 | 0.000180 | 0.000158 |
