You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004739_01676
You are here: Home > Sequence: MGYG000004739_01676
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Actinomyces sp000220835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; Actinomyces sp000220835 | |||||||||||
| CAZyme ID | MGYG000004739_01676 | |||||||||||
| CAZy Family | GT87 | |||||||||||
| CAZyme Description | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1077; End: 2459 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT87 | 83 | 294 | 4.6e-58 | 0.9134199134199135 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13375 | pimE | 2.01e-28 | 52 | 380 | 46 | 337 | mannosyltransferase; Provisional |
| pfam09594 | GT87 | 1.48e-19 | 83 | 262 | 1 | 177 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMD88459.1 | 2.04e-203 | 27 | 436 | 3 | 394 |
| CED91729.1 | 7.66e-189 | 14 | 444 | 3 | 411 |
| VEG28296.1 | 7.61e-185 | 28 | 433 | 4 | 405 |
| QHO91881.1 | 6.25e-183 | 14 | 444 | 3 | 417 |
| QQC21714.1 | 8.22e-168 | 8 | 437 | 34 | 458 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WMZ8 | 9.28e-40 | 29 | 451 | 23 | 423 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2236 PE=3 SV=1 |
| P9WMZ9 | 9.28e-40 | 29 | 451 | 23 | 423 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv2181 PE=1 SV=1 |
| A0R036 | 8.22e-39 | 30 | 432 | 26 | 411 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=MSMEG_4247 PE=1 SV=1 |
| A0R2K8 | 2.20e-23 | 24 | 369 | 28 | 338 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol pentamannoside mannosyltransferase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=pimE PE=1 SV=2 |
| P9WN00 | 1.62e-19 | 30 | 369 | 46 | 351 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol pentamannoside mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pimE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999985 | 0.000009 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
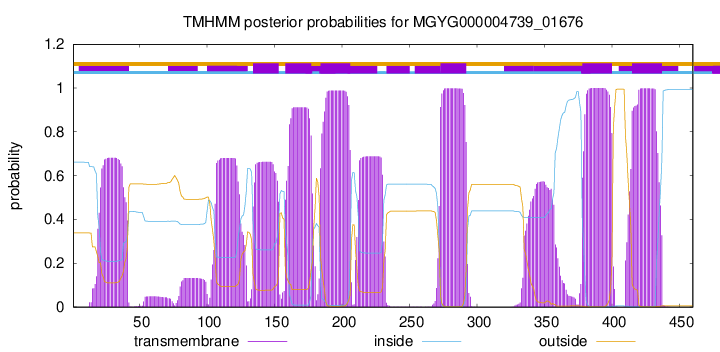
| start | end |
|---|---|
| 134 | 153 |
| 158 | 177 |
| 184 | 206 |
| 273 | 292 |
| 378 | 400 |
| 415 | 437 |
