You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004748_01038
You are here: Home > Sequence: MGYG000004748_01038
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000004748_01038 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | Unsaturated rhamnogalacturonyl hydrolase YteR | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30051; End: 31151 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 51 | 362 | 4.2e-85 | 0.9668674698795181 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 5.01e-89 | 38 | 364 | 5 | 342 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 1.98e-51 | 55 | 364 | 35 | 356 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH57430.1 | 4.46e-256 | 1 | 365 | 1 | 365 |
| QUT45004.1 | 2.11e-251 | 1 | 366 | 1 | 366 |
| QRQ49255.1 | 1.22e-250 | 1 | 366 | 1 | 366 |
| QDO70976.1 | 3.16e-239 | 1 | 366 | 1 | 365 |
| QRQ48187.1 | 1.33e-224 | 1 | 363 | 1 | 363 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CE7_A | 6.16e-74 | 15 | 364 | 7 | 369 | Crystalstructure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_B Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_C Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans] |
| 5NOA_A | 1.60e-55 | 1 | 366 | 1 | 376 | PolysaccharideLyase BACCELL_00875 [Bacteroides thetaiotaomicron] |
| 4Q88_A | 1.15e-47 | 29 | 366 | 14 | 359 | ChainA, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482],4Q88_B Chain B, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WF04 | 3.73e-77 | 33 | 361 | 59 | 408 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_28 PE=1 SV=1 |
| L7P9J4 | 2.07e-73 | 7 | 364 | 6 | 376 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Nonlabens ulvanivorans OX=906888 GN=IL45_01505 PE=1 SV=1 |
| T2KPL9 | 4.58e-72 | 53 | 366 | 55 | 377 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22220 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
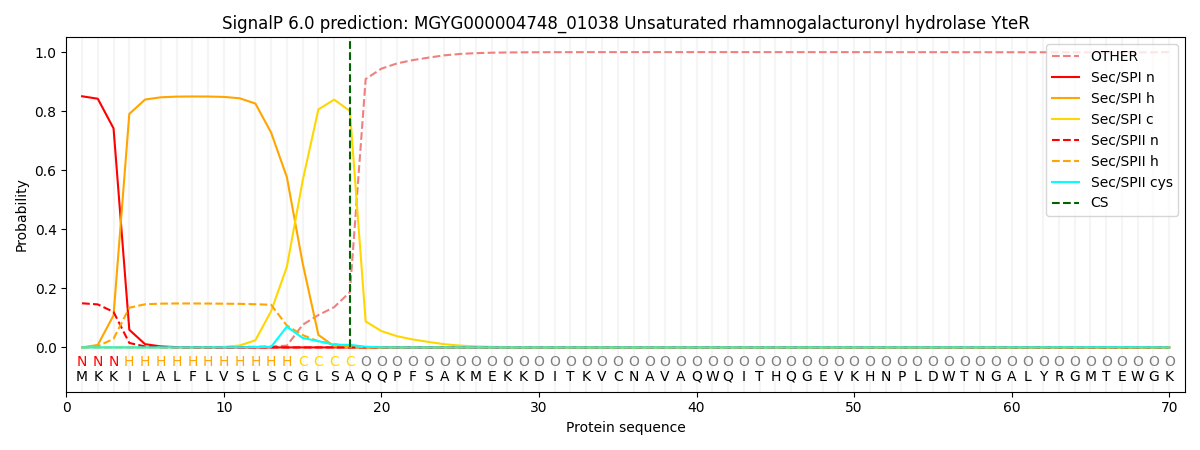
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000867 | 0.843399 | 0.154943 | 0.000292 | 0.000253 | 0.000230 |
