You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004752_00966
You are here: Home > Sequence: MGYG000004752_00966
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
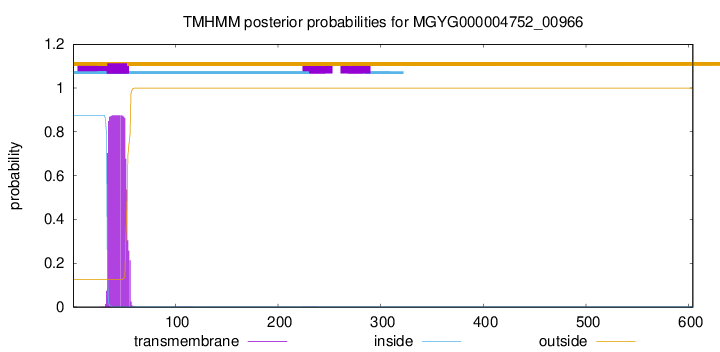
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Roseburia; | |||||||||||
| CAZyme ID | MGYG000004752_00966 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1669; End: 3483 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 344 | 586 | 3.4e-78 | 0.46435845213849286 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14251 | PL-6 | 2.67e-04 | 375 | 537 | 12 | 166 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam13229 | Beta_helix | 0.004 | 431 | 551 | 3 | 124 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam14592 | Chondroitinas_B | 0.004 | 377 | 550 | 14 | 214 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
| pfam18998 | Flg_new_2 | 0.004 | 282 | 336 | 18 | 72 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDS97815.1 | 1.07e-119 | 63 | 585 | 340 | 854 |
| AIY03209.1 | 1.99e-113 | 55 | 585 | 314 | 835 |
| QLL50162.1 | 8.39e-85 | 64 | 585 | 141 | 650 |
| QLL52630.1 | 8.39e-85 | 64 | 585 | 141 | 650 |
| ALA27928.1 | 8.39e-85 | 64 | 585 | 141 | 650 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 2.69e-33 | 345 | 585 | 11 | 273 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 6KQS_A | 1.96e-23 | 345 | 585 | 247 | 503 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 6KQT_A | 2.60e-23 | 345 | 585 | 247 | 503 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 5GQC_A | 6.45e-21 | 345 | 585 | 20 | 311 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 1.49e-19 | 345 | 585 | 16 | 315 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
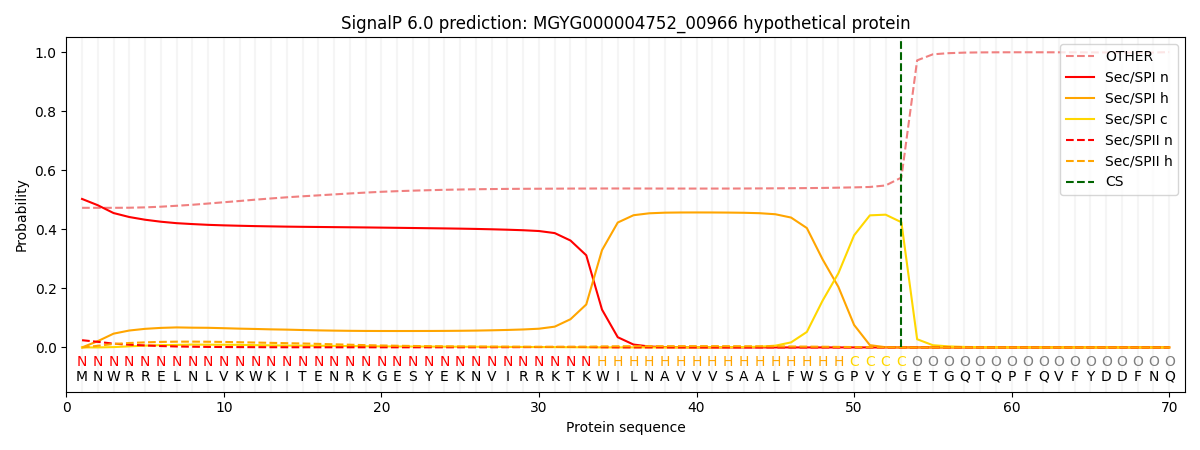
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.480990 | 0.488980 | 0.028767 | 0.000523 | 0.000310 | 0.000422 |