You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004754_00234
You are here: Home > Sequence: MGYG000004754_00234
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Elizabethkingia bruuniana | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Flavobacteriales; Weeksellaceae; Elizabethkingia; Elizabethkingia bruuniana | |||||||||||
| CAZyme ID | MGYG000004754_00234 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 266087; End: 267658 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 399 | 520 | 8.8e-19 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.53e-14 | 398 | 520 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 0.001 | 396 | 476 | 14 | 92 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATL43245.1 | 0.0 | 1 | 523 | 1 | 523 |
| QDZ63052.1 | 0.0 | 1 | 523 | 1 | 523 |
| QQN57758.1 | 0.0 | 1 | 523 | 1 | 523 |
| AQX84323.1 | 0.0 | 15 | 523 | 1 | 509 |
| QQD13103.1 | 1.24e-178 | 9 | 523 | 7 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Z2D_A | 8.21e-31 | 50 | 361 | 21 | 338 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
| 6Z2O_A | 8.35e-31 | 50 | 361 | 22 | 339 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835],6Z2Q_A Chain A, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
| 6Z2P_A | 1.23e-28 | 50 | 361 | 22 | 339 | ChainA, O-glycan protease [Akkermansia muciniphila ATCC BAA-835] |
| 3F2Z_A | 3.79e-11 | 386 | 505 | 3 | 128 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
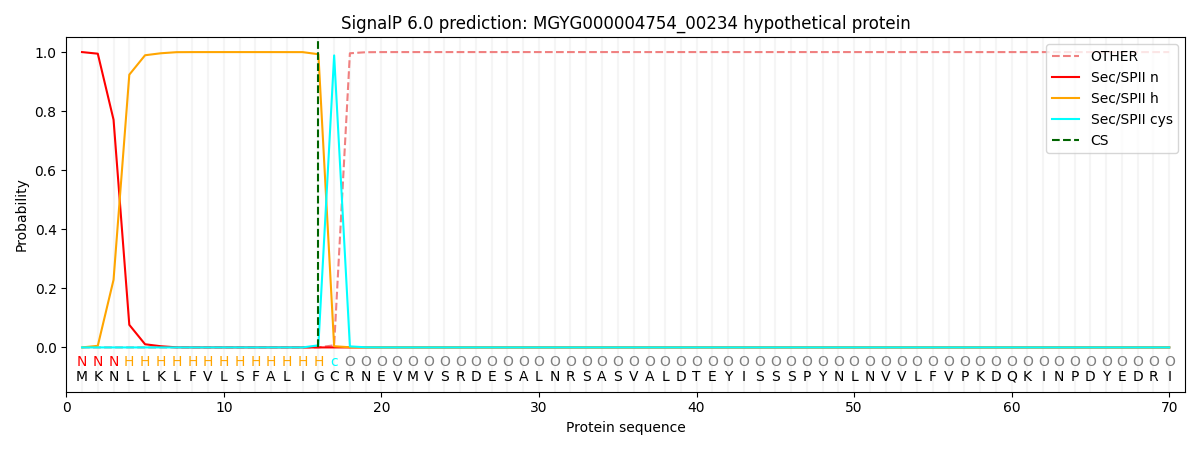
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000009 | 1.000043 | 0.000000 | 0.000000 | 0.000000 |
