You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004758_01546
You are here: Home > Sequence: MGYG000004758_01546
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900553155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900553155 | |||||||||||
| CAZyme ID | MGYG000004758_01546 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10006; End: 12546 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 522 | 742 | 1.2e-16 | 0.7837837837837838 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 5.06e-10 | 46 | 160 | 17 | 145 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam00704 | Glyco_hydro_18 | 4.36e-09 | 498 | 732 | 40 | 307 | Glycosyl hydrolases family 18. |
| pfam13385 | Laminin_G_3 | 2.81e-06 | 302 | 455 | 1 | 143 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| smart00636 | Glyco_18 | 4.48e-06 | 501 | 732 | 46 | 334 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ12875.1 | 2.64e-223 | 19 | 792 | 23 | 797 |
| QUT75436.1 | 5.57e-184 | 4 | 828 | 6 | 837 |
| QDO70362.1 | 2.16e-131 | 11 | 845 | 11 | 861 |
| AHF11971.1 | 1.37e-116 | 11 | 843 | 13 | 849 |
| BCI61676.1 | 4.03e-116 | 16 | 845 | 18 | 853 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
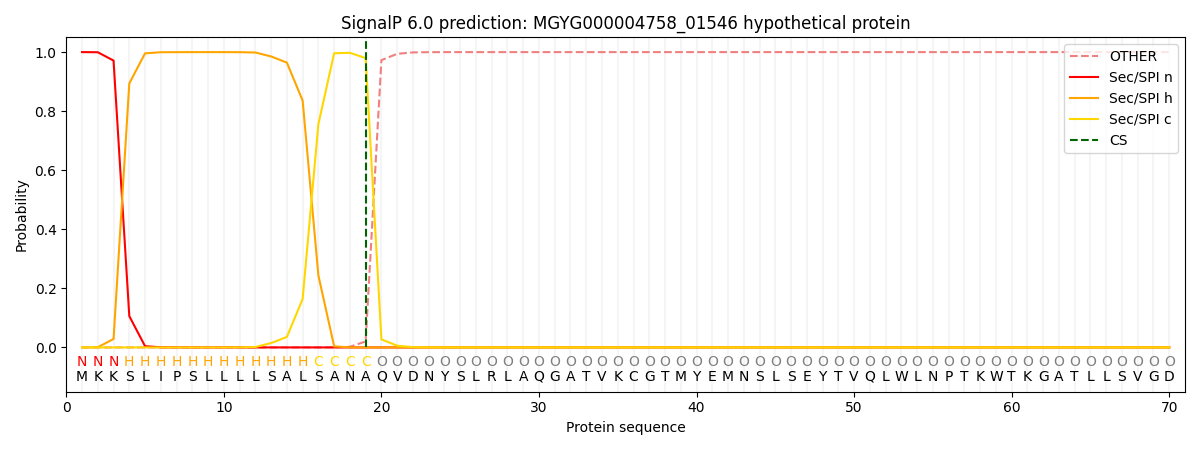
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000303 | 0.998971 | 0.000193 | 0.000188 | 0.000173 | 0.000157 |
