You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004763_00271
You are here: Home > Sequence: MGYG000004763_00271
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp900291465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp900291465 | |||||||||||
| CAZyme ID | MGYG000004763_00271 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34477; End: 37731 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 358 | 810 | 5.6e-114 | 0.49393203883495146 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 39 | 881 | 5 | 866 | alpha-L-rhamnosidase. |
| cd00057 | FA58C | 2.69e-04 | 220 | 294 | 47 | 127 | Substituted updates: Jan 31, 2002 |
| cd04081 | CBM35_galactosidase-like | 0.005 | 999 | 1040 | 69 | 117 | Carbohydrate Binding Module family 35 (CBM35); appended mainly to enzymes that bind alpha-D-galactose (CBM35-Gal), including glycoside hydrolase (GH) families GH27 and GH43. This family includes carbohydrate binding module family 35 (CBM35); these are non-catalytic carbohydrate binding domains that are appended mainly to enzymes that bind alpha-D-galactose (CBM35-Gal), including glycoside hydrolase (GH) families GH27 and GH43. Examples of proteins which contain CBM35s belonging to this family includes the CBM35 of an exo-beta-1,3-galactanase from Phanerochaete chrysosporium 9 (Pc1,3Gal43A) which is appended to a GH43 domain, and the CBM35 domain of two bifunctional proteins with beta-L-arabinopyranosidase/alpha-D-galactopyranosidase activities from Fusarium oxysporum 12S, Foap1 and Foap2 (Fo/AP1 and Fo/AP2), that are appended to GH27 domains. CBM35s are unique in that they display conserved specificity through extensive sequence similarity but divergent function through their appended catalytic modules. They are known to bind alpha-D-galactose (Gal), mannan (Man), xylan, glucuronic acid (GlcA), a beta-polymer of mannose, and possibly glucans, forming four subfamilies based on general ligand specificities (galacto, urono, manno, and gluco configurations). Some CBM35s bind their ligands in a calcium-dependent manner. In contrast to most CBMs that are generally rigid proteins, CBM35 undergoes significant conformational change upon ligand binding. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates, while family GH27 includes alpha-galactosidases, alpha-N-acetylgalactosaminidases, and isomaltodextranases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAO76093.1 | 0.0 | 3 | 1080 | 2 | 1098 |
| ALJ42141.1 | 0.0 | 3 | 1080 | 2 | 1098 |
| QQA06590.1 | 0.0 | 3 | 1080 | 2 | 1098 |
| QMW88010.1 | 0.0 | 3 | 1080 | 2 | 1098 |
| QUT70999.1 | 0.0 | 3 | 1082 | 2 | 1100 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 0.0 | 3 | 1080 | 2 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 0.0 | 3 | 1080 | 2 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6Q2F_A | 3.09e-121 | 17 | 1078 | 30 | 1136 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 5.11e-96 | 29 | 924 | 23 | 751 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
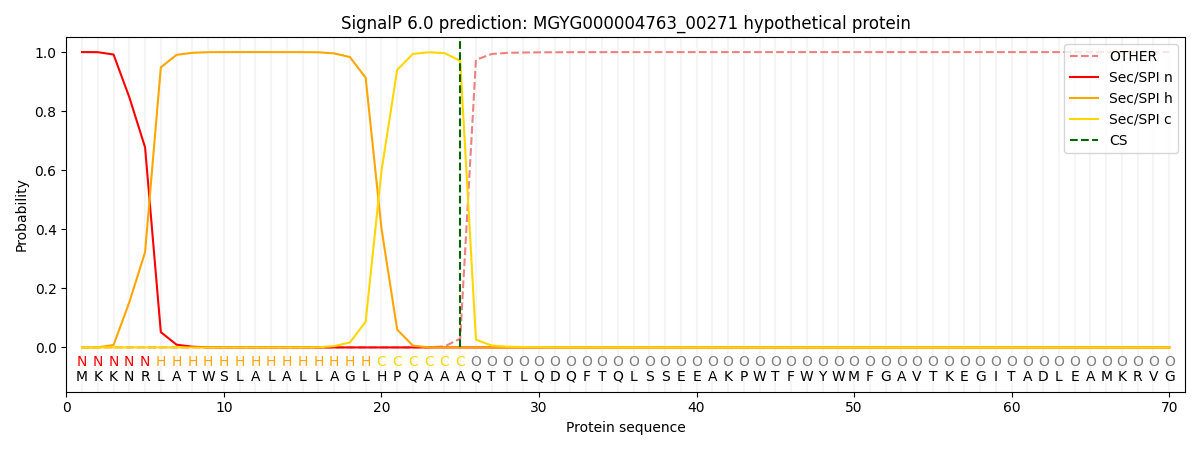
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000305 | 0.998999 | 0.000230 | 0.000164 | 0.000152 | 0.000144 |
