You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004763_01899
You are here: Home > Sequence: MGYG000004763_01899
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp900291465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp900291465 | |||||||||||
| CAZyme ID | MGYG000004763_01899 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2979; End: 4496 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 203 | 482 | 1.3e-40 | 0.9513888888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02671 | PLN02671 | 3.36e-14 | 200 | 478 | 58 | 337 | pectinesterase |
| COG4677 | PemB | 1.25e-13 | 184 | 480 | 65 | 390 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02682 | PLN02682 | 1.02e-12 | 202 | 478 | 70 | 347 | pectinesterase family protein |
| PLN02304 | PLN02304 | 2.89e-12 | 200 | 487 | 74 | 363 | probable pectinesterase |
| PLN02773 | PLN02773 | 1.64e-10 | 203 | 485 | 7 | 286 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJR79100.1 | 3.16e-289 | 9 | 502 | 2 | 498 |
| QJR66174.1 | 3.16e-289 | 9 | 502 | 2 | 498 |
| AII67872.1 | 3.16e-289 | 9 | 502 | 2 | 498 |
| ALA76100.1 | 3.16e-289 | 9 | 502 | 2 | 498 |
| QJR57681.1 | 3.16e-289 | 9 | 502 | 2 | 498 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O23038 | 1.97e-11 | 197 | 487 | 82 | 377 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q9FM79 | 4.24e-09 | 191 | 482 | 66 | 364 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9LSP1 | 4.73e-08 | 174 | 365 | 10 | 201 | Probable pectinesterase 67 OS=Arabidopsis thaliana OX=3702 GN=PME67 PE=2 SV=1 |
| Q9ZQA3 | 5.99e-08 | 203 | 449 | 91 | 336 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q9LVQ0 | 4.09e-07 | 198 | 455 | 2 | 263 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
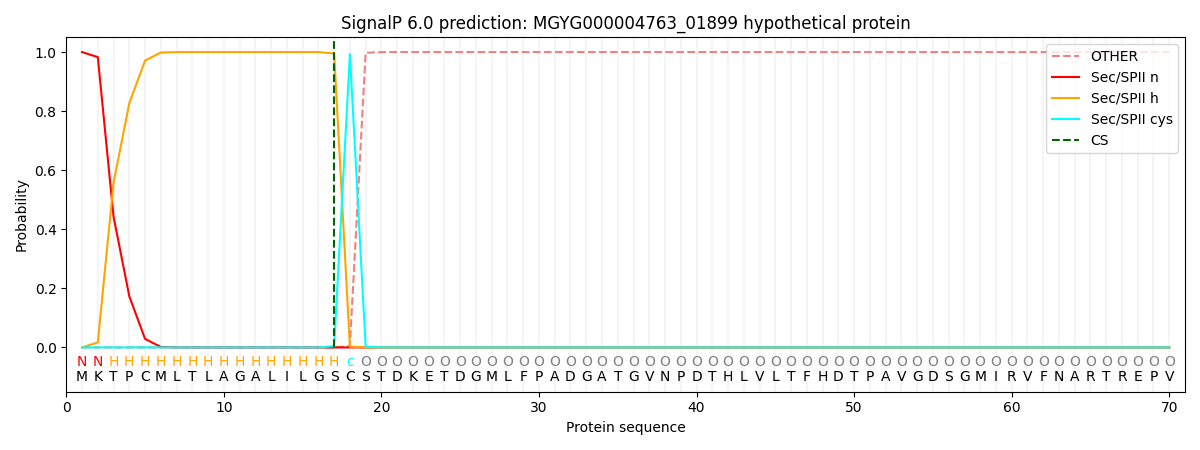
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000075 | 0.000000 | 0.000000 | 0.000000 |
