You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004773_00402
You are here: Home > Sequence: MGYG000004773_00402
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Succinivibrio sp902776275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Succinivibrionaceae; Succinivibrio; Succinivibrio sp902776275 | |||||||||||
| CAZyme ID | MGYG000004773_00402 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47524; End: 48180 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 7 | 216 | 1.5e-67 | 0.7162629757785467 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 3.07e-30 | 1 | 184 | 21 | 187 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 4.87e-04 | 9 | 183 | 26 | 170 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADT88760.1 | 6.97e-95 | 1 | 218 | 61 | 279 |
| AMF93025.1 | 6.97e-95 | 1 | 218 | 61 | 279 |
| QDC94579.1 | 6.97e-95 | 1 | 218 | 61 | 279 |
| BAG69482.2 | 5.71e-94 | 1 | 218 | 63 | 281 |
| QIL87027.1 | 7.86e-94 | 1 | 218 | 61 | 279 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PZ9_A | 1.11e-72 | 1 | 218 | 37 | 269 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 4.56e-70 | 1 | 218 | 23 | 255 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 4QP0_A | 1.23e-42 | 9 | 218 | 37 | 257 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3ZIZ_A | 6.30e-32 | 6 | 218 | 46 | 252 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
| 1RH9_A | 2.15e-29 | 5 | 218 | 40 | 252 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SG95 | 4.49e-36 | 5 | 184 | 63 | 242 | Putative mannan endo-1,4-beta-mannosidase 4 OS=Arabidopsis thaliana OX=3702 GN=MAN4 PE=3 SV=1 |
| Q9LW44 | 1.26e-34 | 5 | 184 | 63 | 242 | Putative mannan endo-1,4-beta-mannosidase P OS=Arabidopsis thaliana OX=3702 GN=MANP PE=5 SV=3 |
| O48540 | 1.52e-33 | 9 | 184 | 65 | 239 | Mannan endo-1,4-beta-mannosidase 1 OS=Solanum lycopersicum OX=4081 GN=MAN1 PE=1 SV=2 |
| Q5W6G0 | 2.71e-33 | 5 | 184 | 112 | 289 | Putative mannan endo-1,4-beta-mannosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN5 PE=2 SV=2 |
| Q0IQJ7 | 3.84e-33 | 5 | 218 | 35 | 252 | Mannan endo-1,4-beta-mannosidase 8 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN8 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
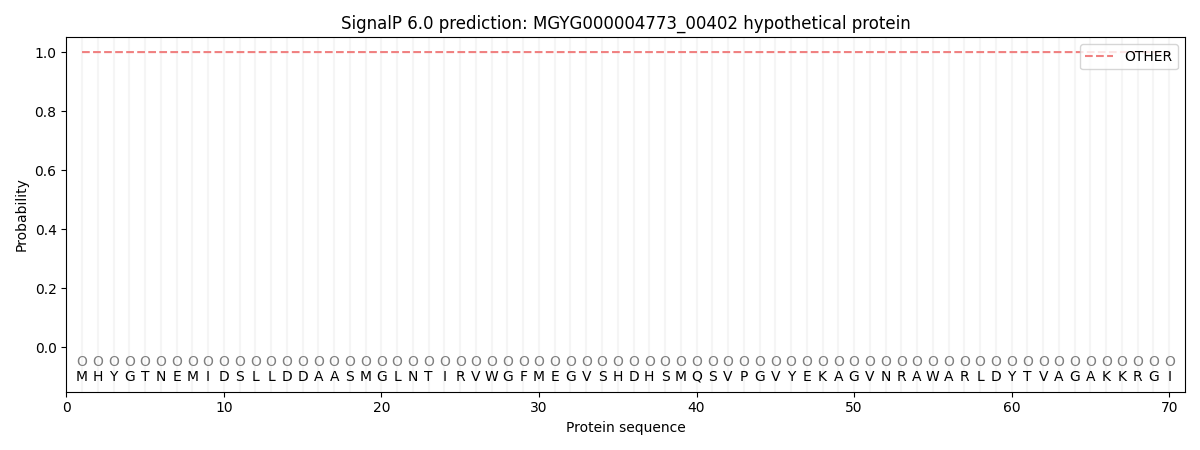
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
