You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004797_01532
You are here: Home > Sequence: MGYG000004797_01532
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sartorii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sartorii | |||||||||||
| CAZyme ID | MGYG000004797_01532 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42269; End: 43789 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 199 | 481 | 1.9e-45 | 0.9618055555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02480 | PLN02480 | 2.17e-14 | 187 | 498 | 32 | 340 | Probable pectinesterase |
| PLN02682 | PLN02682 | 3.93e-14 | 198 | 475 | 70 | 348 | pectinesterase family protein |
| PLN02671 | PLN02671 | 7.72e-14 | 181 | 478 | 44 | 341 | pectinesterase |
| PLN02773 | PLN02773 | 3.39e-13 | 199 | 481 | 7 | 288 | pectinesterase |
| PLN02304 | PLN02304 | 3.70e-13 | 191 | 484 | 70 | 364 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT59621.1 | 0.0 | 1 | 503 | 1 | 503 |
| ABR39479.1 | 0.0 | 1 | 503 | 10 | 512 |
| QQY40500.1 | 0.0 | 1 | 503 | 1 | 503 |
| QQY44776.1 | 0.0 | 1 | 503 | 1 | 503 |
| QJR79100.1 | 0.0 | 1 | 503 | 1 | 503 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O23038 | 6.28e-12 | 199 | 484 | 91 | 378 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q9FM79 | 2.47e-11 | 199 | 478 | 82 | 362 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9ZQA3 | 2.06e-10 | 182 | 484 | 75 | 375 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q9LSP1 | 1.51e-08 | 188 | 501 | 29 | 344 | Probable pectinesterase 67 OS=Arabidopsis thaliana OX=3702 GN=PME67 PE=2 SV=1 |
| Q9LY18 | 3.85e-08 | 199 | 505 | 67 | 357 | Probable pectinesterase 49 OS=Arabidopsis thaliana OX=3702 GN=PME49 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
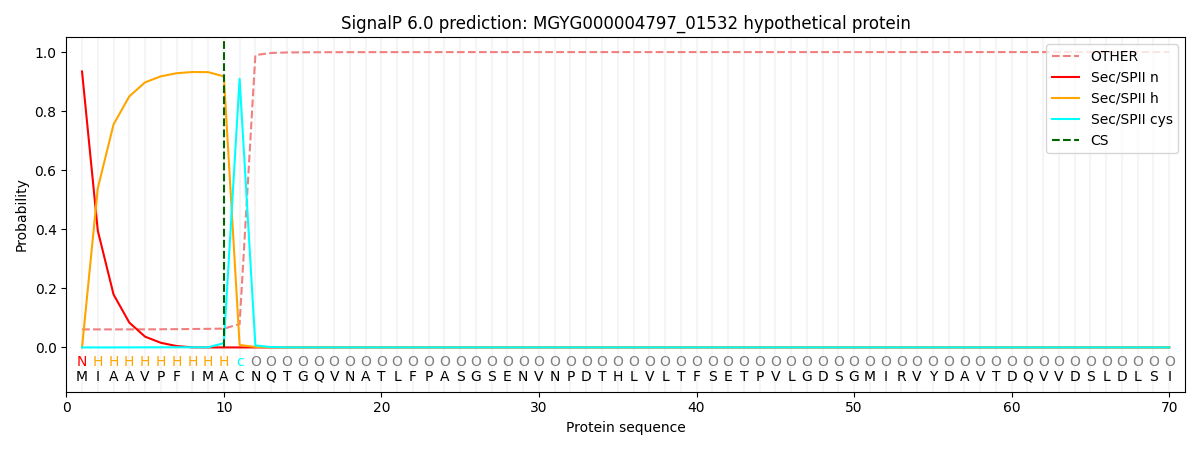
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.061211 | 0.004325 | 0.934467 | 0.000004 | 0.000006 | 0.000006 |
