You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004797_02793
You are here: Home > Sequence: MGYG000004797_02793
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sartorii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sartorii | |||||||||||
| CAZyme ID | MGYG000004797_02793 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20473; End: 22536 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 23 | 210 | 2.4e-16 | 0.979381443298969 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01827 | sialate_O-acetylesterase_like1 | 1.60e-83 | 23 | 214 | 1 | 188 | sialate O-acetylesterase_like family of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 3.79e-28 | 27 | 201 | 1 | 173 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam03629 | SASA | 5.90e-22 | 301 | 576 | 2 | 221 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| cd00229 | SGNH_hydrolase | 7.61e-20 | 25 | 207 | 1 | 182 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd01834 | SGNH_hydrolase_like_2 | 7.92e-13 | 24 | 208 | 3 | 187 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB42485.1 | 3.58e-269 | 21 | 684 | 29 | 692 |
| AHW60353.1 | 7.70e-249 | 20 | 687 | 33 | 703 |
| AWI09525.1 | 2.46e-102 | 220 | 685 | 636 | 1123 |
| QDT62262.1 | 2.19e-97 | 215 | 684 | 945 | 1438 |
| QUT73831.1 | 7.03e-92 | 210 | 687 | 14 | 473 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KMM_A | 2.90e-61 | 222 | 687 | 25 | 634 | ChainA, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306],7KMM_B Chain B, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82450 | 1.75e-40 | 219 | 630 | 25 | 469 | Sialate O-acetylesterase OS=Rattus norvegicus OX=10116 GN=Siae PE=1 SV=2 |
| P70665 | 2.72e-39 | 219 | 630 | 25 | 468 | Sialate O-acetylesterase OS=Mus musculus OX=10090 GN=Siae PE=1 SV=3 |
| Q5RFU0 | 2.36e-36 | 219 | 612 | 25 | 425 | Sialate O-acetylesterase OS=Pongo abelii OX=9601 GN=SIAE PE=2 SV=1 |
| Q9HAT2 | 4.32e-36 | 219 | 612 | 25 | 425 | Sialate O-acetylesterase OS=Homo sapiens OX=9606 GN=SIAE PE=1 SV=1 |
| D5EV35 | 2.14e-28 | 20 | 207 | 271 | 473 | Acetylxylan esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axeA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
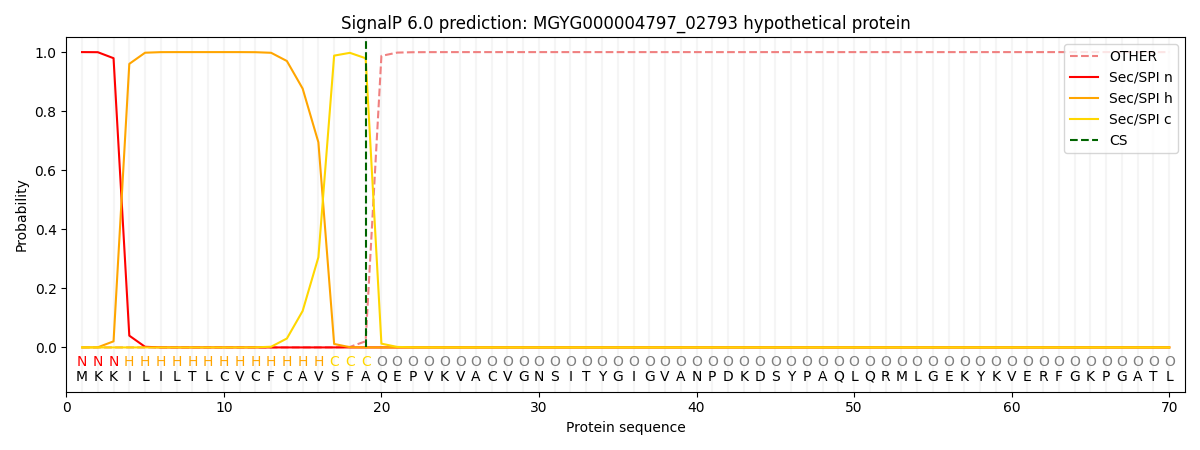
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000225 | 0.999107 | 0.000218 | 0.000144 | 0.000146 | 0.000139 |
