You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004798_01429
You are here: Home > Sequence: MGYG000004798_01429
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
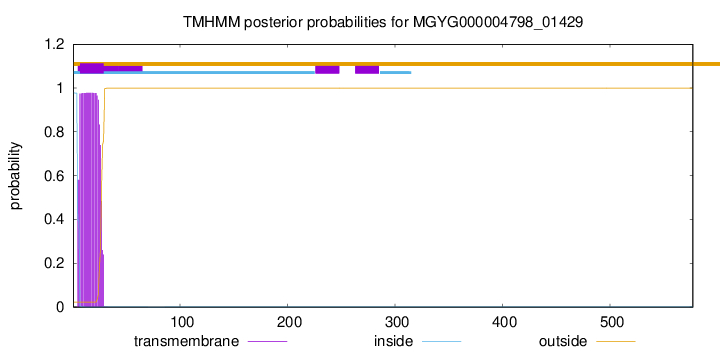
TMHMM annotations
Basic Information help
| Species | Enterococcus_B hirae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_B; Enterococcus_B hirae | |||||||||||
| CAZyme ID | MGYG000004798_01429 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34807; End: 36540 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 41 | 333 | 2.2e-28 | 0.9831081081081081 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3469 | Chi1 | 4.52e-143 | 16 | 335 | 4 | 325 | Chitinase [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 1.60e-89 | 39 | 338 | 1 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| pfam00704 | Glyco_hydro_18 | 6.41e-15 | 42 | 328 | 3 | 271 | Glycosyl hydrolases family 18. |
| cd00598 | GH18_chitinase-like | 7.10e-13 | 42 | 223 | 2 | 177 | The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. |
| cd12214 | ChiA1_BD | 1.28e-12 | 532 | 576 | 1 | 45 | chitin-binding domain of Chi A1-like proteins. This group contains proteins related to the chitin binding domain of chitinase A1 (ChiA1) of Bacillus circulans WL-12. Glycosidase ChiA1 hydrolyzes chitin and is comprised of several domains: the C-terminal chitin binding domain, an N-terminal and catalytic domain, and 2 fibronectin type III-like domains. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. Bacillus circulans WL-12 ChiA1 facilitates invasion of fungal cell walls. The ChiAi chitin binding domain is required for the specific recognition of insoluble chitin. although topologically and structurally related, ChiA1 lacks the characteristic aromatic residues of Erwinia chrysanthemi endoglucanase Z (CBD(EGZ)). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV81623.1 | 0.0 | 1 | 577 | 1 | 577 |
| QKX70210.1 | 0.0 | 1 | 577 | 1 | 577 |
| VEE77387.1 | 0.0 | 1 | 577 | 1 | 577 |
| QKX67638.1 | 0.0 | 1 | 577 | 1 | 577 |
| AND73199.1 | 0.0 | 1 | 577 | 1 | 577 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4AXN_A | 1.93e-128 | 25 | 340 | 11 | 328 | Hallmarksof processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases [Serratia marcescens],4AXN_B Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases [Serratia marcescens] |
| 3IAN_A | 3.29e-119 | 39 | 339 | 5 | 310 | ChainA, Chitinase [Lactococcus lactis subsp. lactis] |
| 4W5Z_A | 6.04e-112 | 40 | 344 | 32 | 343 | Highresolution crystal structure of catalytic domain of Chitinase 60 from psychrophilic bacteria Moritella marina. [Moritella marina] |
| 4HMC_A | 4.77e-110 | 40 | 549 | 10 | 504 | Crystalstructure of cold-adapted chitinase from Moritella marina [Moritella marina],4HMD_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) [Moritella marina],4HME_A Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 [Moritella marina] |
| 4MB3_A | 1.34e-109 | 40 | 549 | 10 | 504 | Crystalstructure of E153Q mutant of cold-adapted chitinase from Moritella marina [Moritella marina],4MB4_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 [Moritella marina],4MB5_A Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 [Moritella marina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q838S2 | 1.57e-132 | 1 | 340 | 1 | 348 | Chitinase OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0361 PE=1 SV=1 |
| P20533 | 1.64e-41 | 114 | 573 | 158 | 696 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P50899 | 5.01e-25 | 335 | 527 | 688 | 887 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
| A5FB63 | 1.01e-23 | 39 | 343 | 1142 | 1484 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| P50401 | 3.86e-17 | 343 | 477 | 476 | 616 | Exoglucanase A OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
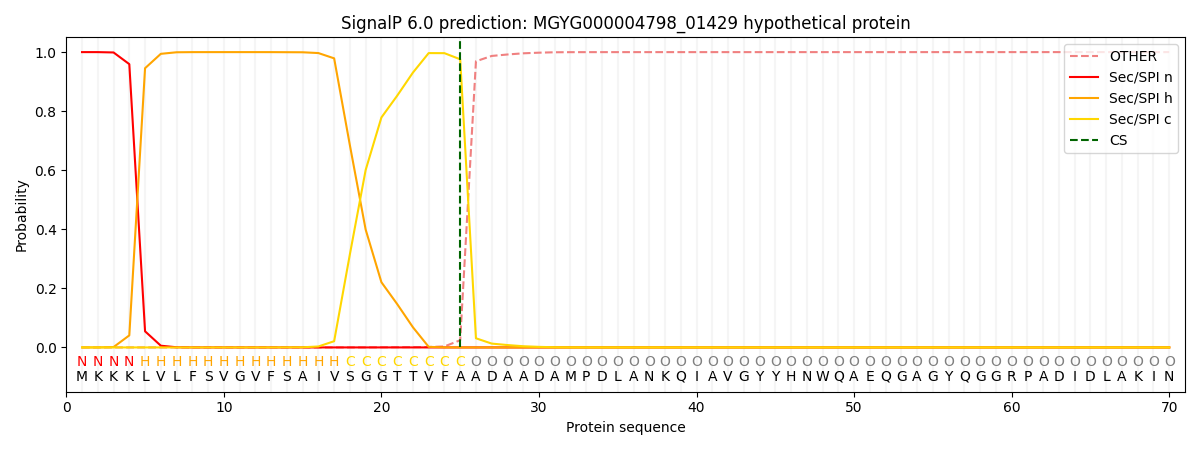
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.998939 | 0.000182 | 0.000194 | 0.000181 | 0.000155 |