You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004823_00561
You are here: Home > Sequence: MGYG000004823_00561
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp900765575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp900765575 | |||||||||||
| CAZyme ID | MGYG000004823_00561 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8970; End: 12113 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH127 | 35 | 566 | 2.9e-207 | 0.9980916030534351 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 3.12e-177 | 35 | 566 | 2 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 4.56e-171 | 24 | 644 | 6 | 588 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| cd20170 | Peptidase_M90-like | 2.02e-04 | 944 | 1041 | 117 | 209 | uncharacterized M90 peptidase family-like proteins. This subfamily contains uncharacterized M90 peptidase-like domains, similar to the Mlc Titration Factor A (MtfA) peptidase from Escherichia coli, also known as the YeeI gene product, which is involved in the control of the glucose-phosphotransferase sensory and regulatory system by inactivation of the repressor Mlc (making large colonies). E. coli MtfA has been shown to have aminopeptidase activity with the presence of a single zinc ion in the active site ligated by two histidines in an HEXXH motif. MtfA is related to the catalytic domain of the anthrax lethal factor and the Mop protein involved in the virulence of Vibrio cholerae; although sequence similarity is low, conservation is observed in the overall structure as well as in the residues around the active site. |
| cd20493 | M34_ATLF_C-like | 2.89e-04 | 944 | 998 | 121 | 178 | C-terminal catalytically active domain of anthrax toxin lethal factor and similar domains; belongs to peptidase family M34. This subfamily includes the C-terminal catalytic domain of anthrax toxin lethal factor (ATLF; EC 3.4.24.83). ATLF and edema factor are enzyme components of anthrax toxin and are carried into the cell by a third component, the protective antigen (PA). ATLF is secreted by Bacillus anthracis to promote disease virulence through disruption of host signaling pathways. ATLF belongs to peptidase family M34 and has the hallmark metalloprotease motif HEXXH motif where the two His residues bind a single zinc atom, and the Glu has a catalytic role. ATLF is a highly selective protease whose major substrates are mitogen-activated protein kinase kinases (MKKs). MKKs are cleaved by ATLF near their N-termini, removing the docking sequence for the downstream cognate mitogen-activated protein kinase. Preferred amino acids around the cleavage site can be denoted BBBBxHxH, in which B denotes Arg or Lys, H denotes a hydrophobic amino acid, and x is any amino acid. At its N-terminus, ATLF has a related PABD domain which lacks the hallmark metalloprotease motif HEXXH. This subfamily also includes Bacillus thuringiensis Vip2Ac-like_2 which belongs to the Vip family of proteins that are secreted during the vegetative growth phase. |
| cd20184 | M34_peptidase_like | 0.001 | 947 | 1040 | 56 | 131 | uncharacterized subfamily of peptidase family M34. Peptidase family M34 (also known as the anthrax lethal factor family) includes the C-terminal catalytic domain of anthrax lethal factor (ATLF, EC 3.4.24.83), and the N-terminal protective antigen-binding domains (PABDs) of ATLF and edema factor (EF). ATLF and EF are enzyme components of anthrax toxin and are carried into the cell by a third component, the protective antigen (PA). ATLF is a highly selective protease whose major substrates are mitogen-activated protein kinase kinases (MKKs). At its N-terminus, ATLF has a PABD domain which lacks the hallmark metalloprotease motif HEXXH, and, at its C-terminus, the related catalytic domain has the HEXXH motif where the two His residues bind a single zinc atom, and the Glu has a catalytic role. EF acts as a Ca2+- and calmodulin-dependent adenylyl cyclase that can cause edema when associated with PA; it is comprised of the PABD and an adenylyl cyclase domain. Pro-Pro endopeptidase (PPEP-1; EC 3.4.24.89, also known as Zmp1) is an extracellular metalloprotease that shows a unique specificity for hydrolyzing a Pro-Pro bond and is involved in bacterial adhesion. This uncharacterized subfamily includes proteins which have an N-terminal SLH domain, and proteins which may have an N-terminal IG-like domain; these proteins have the hallmark metalloprotease motif HEXXH motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO71484.1 | 0.0 | 27 | 807 | 31 | 811 |
| QUT89696.1 | 0.0 | 22 | 807 | 35 | 820 |
| ALJ59245.1 | 0.0 | 22 | 807 | 35 | 820 |
| QQY38942.1 | 0.0 | 20 | 807 | 24 | 811 |
| ABR38820.1 | 0.0 | 20 | 807 | 24 | 811 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EX6_A | 9.53e-201 | 27 | 652 | 7 | 634 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4QJY_A | 4.81e-147 | 33 | 644 | 14 | 648 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 4QK0_A | 1.66e-143 | 33 | 644 | 14 | 648 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 5MQO_A | 1.82e-99 | 35 | 642 | 44 | 695 | Glycosidehydrolase BT_1003 [Bacteroides thetaiotaomicron] |
| 3WRE_A | 6.58e-98 | 69 | 640 | 56 | 655 | Thecrystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 [Bifidobacterium longum subsp. longum JCM 1217],3WRG_A The complex structure of HypBA1 with L-arabinose [Bifidobacterium longum subsp. longum JCM 1217] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH8 | 3.60e-97 | 69 | 640 | 56 | 655 | Non-reducing end beta-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
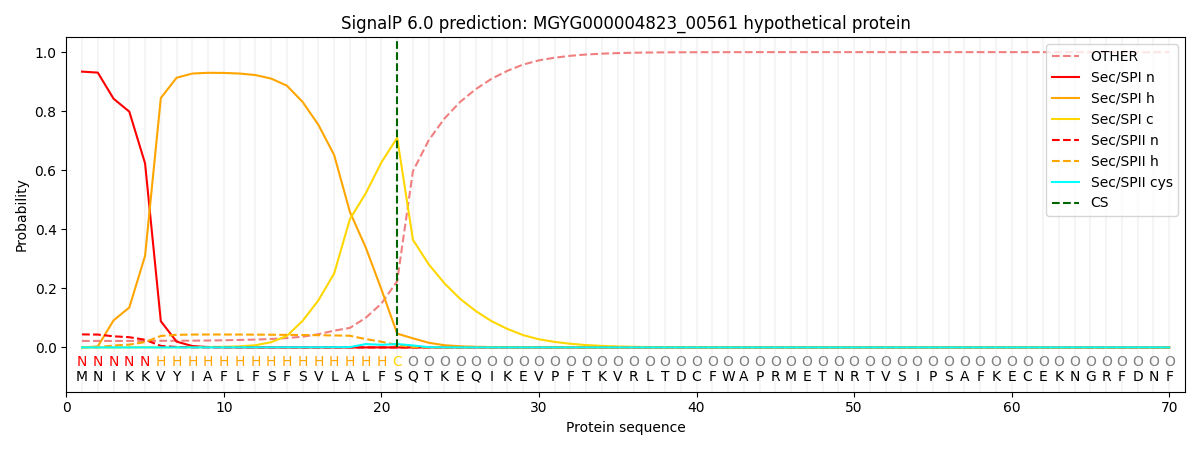
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.024649 | 0.928501 | 0.045976 | 0.000294 | 0.000264 | 0.000270 |
