You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004824_00276
You are here: Home > Sequence: MGYG000004824_00276
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; | |||||||||||
| CAZyme ID | MGYG000004824_00276 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | Putative non-heme bromoperoxidase BpoC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30879; End: 31793 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0596 | MhpC | 3.52e-32 | 30 | 298 | 2 | 282 | Pimeloyl-ACP methyl ester carboxylesterase [Coenzyme transport and metabolism, General function prediction only]. |
| TIGR02427 | protocat_pcaD | 1.61e-28 | 39 | 296 | 1 | 251 | 3-oxoadipate enol-lactonase. Members of this family are 3-oxoadipate enol-lactonase. Note that the substrate is known as 3-oxoadipate enol-lactone, 2-oxo-2,3-dihydrofuran-5-acetate, 4,5-Dihydro-5-oxofuran-2-acetate, and 5-oxo-4,5-dihydrofuran-2-acetate. The enzyme the catalyzes the fourth step in the protocatechuate degradation to beta-ketoadipate and then to succinyl-CoA and acetyl-CoA. 4-hydroxybenzoate, 3-hydroxybenzoate, and vanillate all can be converted in one step to protocatechuate. This enzyme also acts in catechol degradation. In genomes that catabolize both catechol and protocatechuate, two forms of this enzyme may be found. All members of the seed alignment for this model were chosen from within protocatechuate degradation operons of at least three genes of the pathway, from genomes with the complete pathway through beta-ketoadipate. [Energy metabolism, Other] |
| pfam00561 | Abhydrolase_1 | 6.08e-20 | 50 | 284 | 2 | 245 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| PRK14875 | PRK14875 | 4.85e-19 | 15 | 295 | 89 | 368 | acetoin dehydrogenase E2 subunit dihydrolipoyllysine-residue acetyltransferase; Provisional |
| pfam12697 | Abhydrolase_6 | 1.93e-14 | 51 | 288 | 1 | 210 | Alpha/beta hydrolase family. This family contains alpha/beta hydrolase enzymes of diverse specificity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT92203.1 | 3.77e-138 | 20 | 295 | 511 | 785 |
| ALJ62047.1 | 5.33e-138 | 20 | 295 | 511 | 785 |
| QIX26164.1 | 2.25e-09 | 48 | 296 | 33 | 293 |
| QGX99118.1 | 4.01e-09 | 40 | 297 | 19 | 285 |
| QMU58029.1 | 3.09e-07 | 40 | 295 | 19 | 280 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EGN_A | 1.68e-21 | 40 | 295 | 11 | 260 | Est816as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_B Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_C Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_D Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_E Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_F Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_G Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium],5EGN_H Est816 as an N-Acyl homoserine lactone degrading enzyme [uncultured bacterium] |
| 4UHC_A | 1.71e-18 | 40 | 303 | 18 | 276 | Structuralstudies of a thermophilic esterase from Thermogutta terrifontis (Native) [Thermogutta terrifontis],4UHD_A Structural studies of a thermophilic esterase from Thermogutta terrifontis (acetate bound) [Thermogutta terrifontis],4UHE_A Structural studies of a thermophilic esterase from Thermogutta terrifontis (malate bound) [Thermogutta terrifontis],4UHF_A Structural studies of a thermophilic esterase from Thermogutta terrifontis (L37A mutant with butyrate bound) [Thermogutta terrifontis] |
| 4UHH_A | 1.03e-17 | 40 | 301 | 18 | 274 | Structuralstudies of a thermophilic esterase from Thermogutta terrifontis (cacodylate complex) [Thermogutta terrifontis] |
| 3FOB_A | 2.14e-17 | 34 | 295 | 16 | 279 | Crystalstructure of bromoperoxidase from Bacillus anthracis [Bacillus anthracis str. Ames],3FOB_B Crystal structure of bromoperoxidase from Bacillus anthracis [Bacillus anthracis str. Ames],3FOB_C Crystal structure of bromoperoxidase from Bacillus anthracis [Bacillus anthracis str. Ames] |
| 1HKH_A | 2.81e-14 | 34 | 145 | 12 | 124 | unligatedgamma lactamase from an Aureobacterium species [Microbacterium],1HKH_B unligated gamma lactamase from an Aureobacterium species [Microbacterium],1HL7_A Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one [Microbacterium sp.],1HL7_B Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one [Microbacterium sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O06734 | 3.90e-14 | 32 | 294 | 4 | 265 | AB hydrolase superfamily protein YisY OS=Bacillus subtilis (strain 168) OX=224308 GN=yisY PE=3 SV=1 |
| P27747 | 5.21e-14 | 22 | 295 | 108 | 372 | Dihydrolipoyllysine-residue acetyltransferase component of acetoin cleaving system OS=Cupriavidus necator (strain ATCC 17699 / DSM 428 / KCTC 22496 / NCIMB 10442 / H16 / Stanier 337) OX=381666 GN=acoC PE=1 SV=3 |
| P9WNH1 | 4.38e-13 | 40 | 300 | 4 | 262 | Putative non-heme bromoperoxidase BpoC OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=bpoC PE=1 SV=1 |
| P9WNH0 | 4.38e-13 | 40 | 300 | 4 | 262 | Putative non-heme bromoperoxidase BpoC OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=bpoC PE=3 SV=1 |
| A8IAD8 | 4.54e-13 | 40 | 299 | 11 | 264 | Putative carbamate hydrolase RutD OS=Azorhizobium caulinodans (strain ATCC 43989 / DSM 5975 / JCM 20966 / LMG 6465 / NBRC 14845 / NCIMB 13405 / ORS 571) OX=438753 GN=rutD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
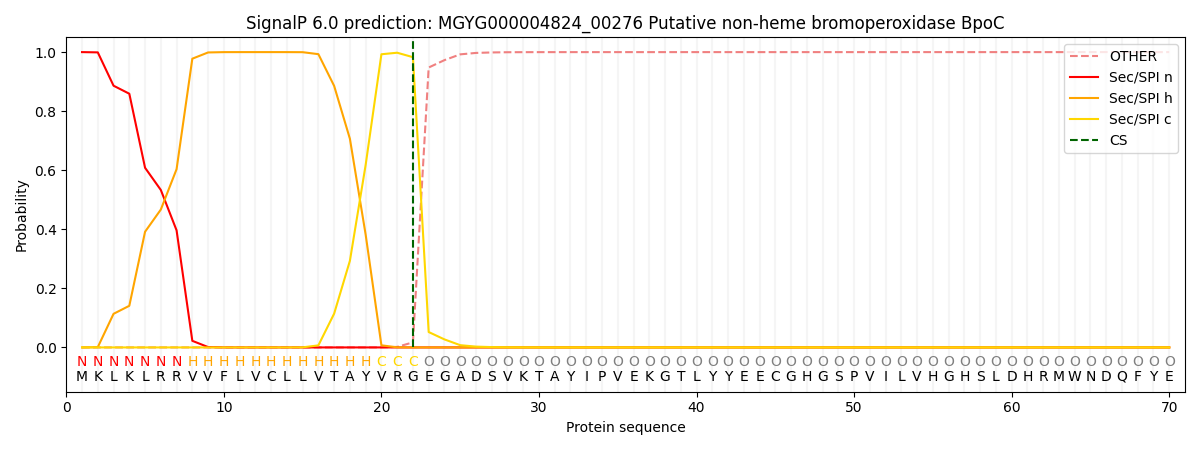
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000211 | 0.999173 | 0.000172 | 0.000153 | 0.000141 | 0.000131 |
