You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004842_01419
You are here: Home > Sequence: MGYG000004842_01419
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
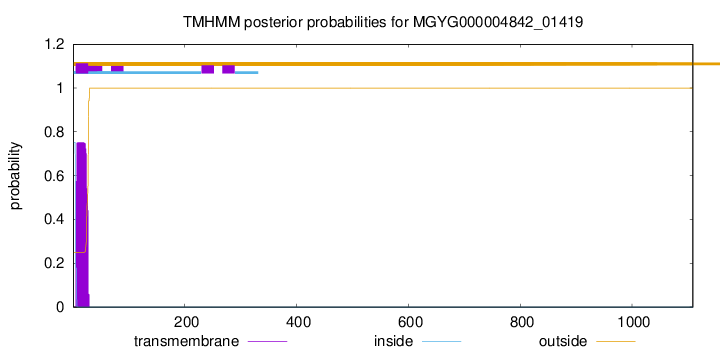
TMHMM annotations
Basic Information help
| Species | Faecalimonas sp900555395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas sp900555395 | |||||||||||
| CAZyme ID | MGYG000004842_01419 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1260; End: 4589 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK14081 | PRK14081 | 2.07e-28 | 557 | 1005 | 6 | 458 | triple tyrosine motif-containing protein; Provisional |
| PRK14081 | PRK14081 | 6.98e-24 | 650 | 1094 | 6 | 458 | triple tyrosine motif-containing protein; Provisional |
| PRK14081 | PRK14081 | 1.62e-23 | 586 | 1001 | 225 | 645 | triple tyrosine motif-containing protein; Provisional |
| COG4193 | LytD | 1.87e-21 | 261 | 439 | 50 | 236 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| PRK14081 | PRK14081 | 1.20e-15 | 763 | 1098 | 21 | 364 | triple tyrosine motif-containing protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOV20091.1 | 3.74e-148 | 88 | 536 | 33 | 480 |
| QEK17141.1 | 3.33e-142 | 71 | 541 | 28 | 500 |
| AWY97555.1 | 3.94e-133 | 94 | 538 | 45 | 490 |
| QQV05342.1 | 1.19e-116 | 1 | 1004 | 5 | 1026 |
| QPS13527.1 | 1.20e-116 | 100 | 1098 | 76 | 1097 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.51e-32 | 194 | 439 | 81 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 3.47e-30 | 171 | 439 | 427 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 4.46e-30 | 171 | 439 | 471 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
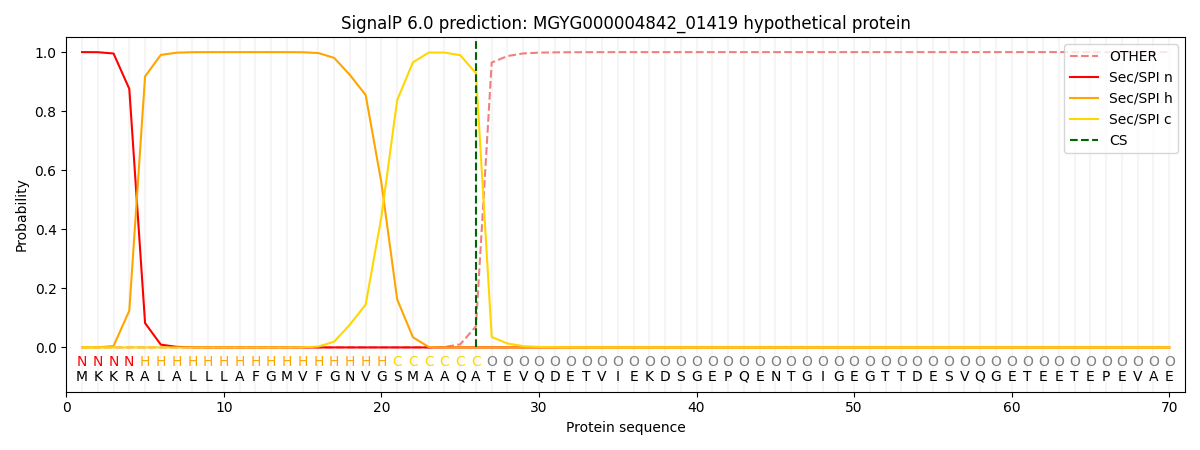
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000508 | 0.998605 | 0.000257 | 0.000211 | 0.000204 | 0.000173 |