You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004845_00755
You are here: Home > Sequence: MGYG000004845_00755
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; UBA4716; | |||||||||||
| CAZyme ID | MGYG000004845_00755 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26624; End: 32794 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 670 | 863 | 4.6e-41 | 0.9836065573770492 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 8.71e-11 | 1853 | 2054 | 569 | 753 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 3.90e-10 | 1938 | 1979 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 1.00e-07 | 1878 | 1919 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 2.78e-07 | 1938 | 2056 | 582 | 694 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 4.86e-07 | 2003 | 2045 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19083.1 | 2.01e-148 | 223 | 2056 | 271 | 2065 |
| QAY65909.1 | 1.35e-42 | 1877 | 2053 | 645 | 821 |
| QEL04270.1 | 9.95e-42 | 604 | 1028 | 1 | 383 |
| QSF47195.1 | 1.45e-41 | 1845 | 2055 | 1073 | 1282 |
| AKP52263.1 | 1.58e-41 | 608 | 1113 | 13 | 462 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 3.45e-16 | 1869 | 2049 | 17 | 192 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 1.20e-15 | 1877 | 2049 | 4 | 171 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 3.95e-29 | 1876 | 2056 | 1281 | 1462 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| Q0CLG7 | 1.44e-24 | 613 | 838 | 17 | 221 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q2UB83 | 1.44e-24 | 614 | 1060 | 18 | 384 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q5B297 | 1.48e-24 | 611 | 1050 | 15 | 352 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| P38536 | 1.67e-24 | 1877 | 2054 | 1681 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
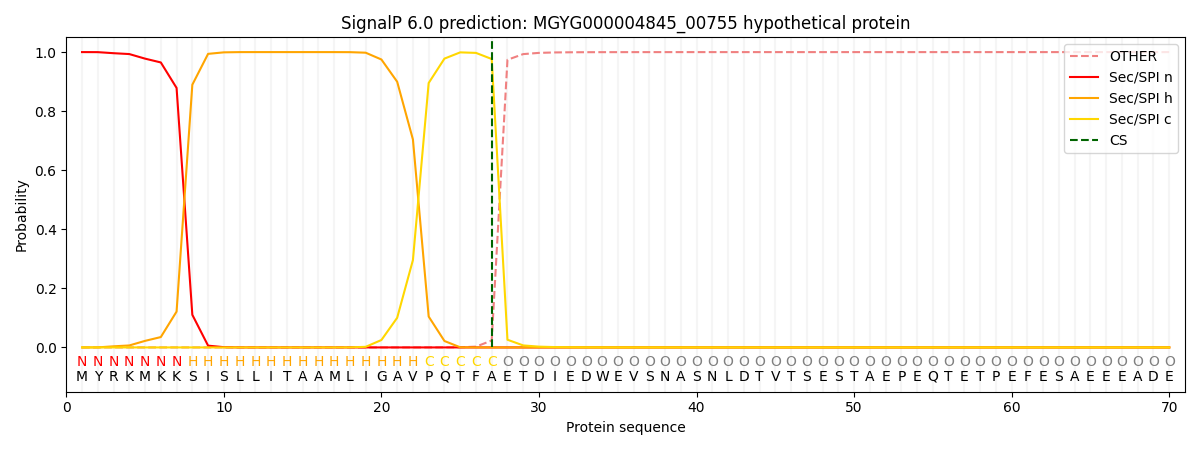
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998929 | 0.000222 | 0.000168 | 0.000168 | 0.000147 |
