You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004845_02099
You are here: Home > Sequence: MGYG000004845_02099
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
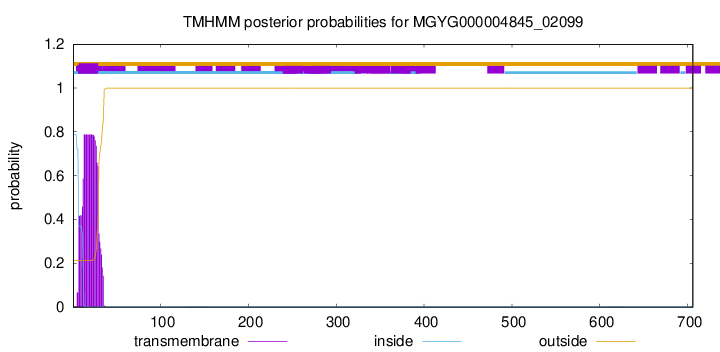
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; UBA4716; | |||||||||||
| CAZyme ID | MGYG000004845_02099 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10763; End: 12883 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 232 | 430 | 1.4e-30 | 0.8811881188118812 |
| CBM13 | 561 | 704 | 1.2e-21 | 0.6914893617021277 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 5.17e-24 | 167 | 472 | 17 | 324 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam14200 | RicinB_lectin_2 | 1.10e-19 | 558 | 641 | 10 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| smart00656 | Amb_all | 2.74e-19 | 243 | 423 | 12 | 181 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 1.06e-11 | 245 | 399 | 32 | 189 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| cd00161 | RICIN | 1.15e-09 | 563 | 702 | 1 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18238.1 | 0.0 | 13 | 705 | 16 | 707 |
| CDM70399.1 | 7.18e-253 | 37 | 705 | 35 | 696 |
| ASR46637.1 | 1.39e-149 | 29 | 528 | 32 | 535 |
| ANY68265.1 | 5.14e-138 | 26 | 528 | 29 | 537 |
| CBL17651.1 | 5.43e-98 | 30 | 668 | 31 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZSC_A | 1.06e-09 | 197 | 398 | 20 | 213 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 1PCL_A | 1.10e-07 | 194 | 474 | 14 | 330 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94449 | 7.79e-23 | 180 | 510 | 35 | 336 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| O34819 | 1.91e-22 | 180 | 510 | 35 | 336 | Pectin lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pelB PE=3 SV=1 |
| Q0CBV0 | 5.86e-15 | 194 | 469 | 42 | 302 | Probable pectate lyase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyB PE=3 SV=1 |
| Q00645 | 1.43e-14 | 194 | 398 | 43 | 235 | Pectate lyase plyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyB PE=1 SV=1 |
| Q2TZY0 | 1.92e-14 | 194 | 398 | 43 | 235 | Probable pectate lyase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
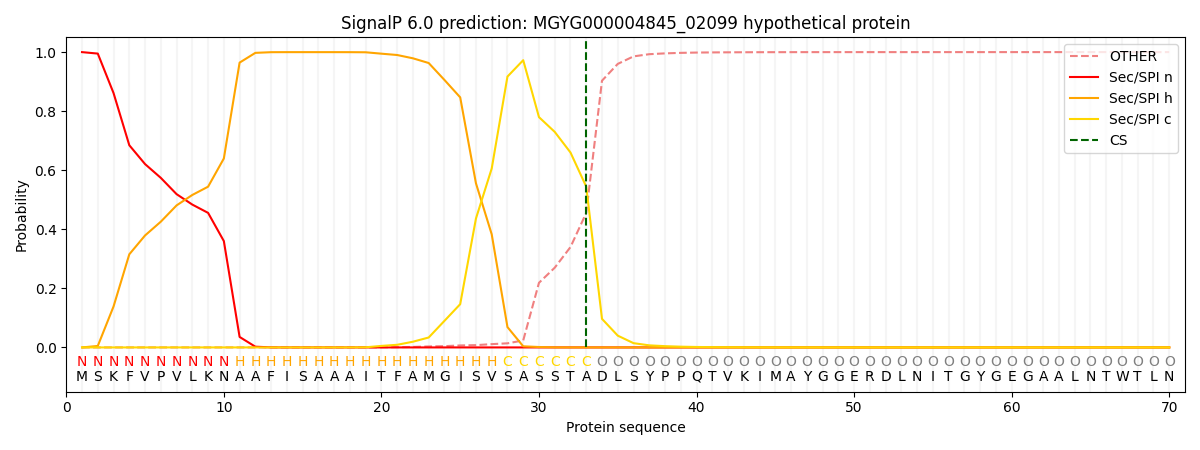
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000397 | 0.998852 | 0.000181 | 0.000192 | 0.000171 | 0.000152 |