You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004845_02428
You are here: Home > Sequence: MGYG000004845_02428
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; UBA4716; | |||||||||||
| CAZyme ID | MGYG000004845_02428 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1312; End: 6240 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 49 | 501 | 8e-136 | 0.9824561403508771 |
| GH43 | 1386 | 1587 | 2e-65 | 0.9904306220095693 |
| CBM62 | 589 | 715 | 1.3e-20 | 0.916030534351145 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08983 | GH43_Bt3655-like | 9.20e-102 | 1372 | 1638 | 1 | 262 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 arabinofuranosidase Bt3655. This glycosyl hydrolase family 43 (GH43)-like family includes the characterized arabinofuranosidases (EC 3.2.1.55): Bacteroides thetaiotaomicron VPI-5482 (Bt3655;BT_3655) and Penicillium chrysogenum 31B Abf43B, as well as Bifidobacterium adolescentis ATCC 15703 beta-xylosidase (EC 3.2.1.37) BAD_1527. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam14587 | Glyco_hydr_30_2 | 9.56e-20 | 56 | 235 | 18 | 195 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 2.01e-18 | 2 | 504 | 4 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| cd08978 | GH_F | 4.43e-17 | 1390 | 1631 | 1 | 244 | Glycosyl hydrolase families 43 and 62 form CAZY clan GH-F. This glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) includes family 43 (GH43) and 62 (GH62). GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH62 includes enzymes characterized as arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. GH62 are also predicted to be inverting enzymes. A common structural feature of both, GH43 and GH62 enzymes, is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam13385 | Laminin_G_3 | 5.47e-12 | 1018 | 1177 | 5 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU23821.1 | 3.18e-139 | 26 | 676 | 22 | 596 |
| QTE67528.1 | 6.75e-130 | 37 | 503 | 31 | 510 |
| AOZ97766.1 | 1.75e-123 | 37 | 504 | 60 | 582 |
| CBK73885.1 | 6.71e-123 | 23 | 714 | 935 | 1634 |
| ADL35667.1 | 2.88e-120 | 37 | 503 | 88 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 3.25e-09 | 86 | 258 | 62 | 221 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A3R0A696 | 7.62e-43 | 1070 | 1613 | 165 | 780 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| A0A401ETL2 | 3.80e-33 | 7 | 459 | 8 | 570 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
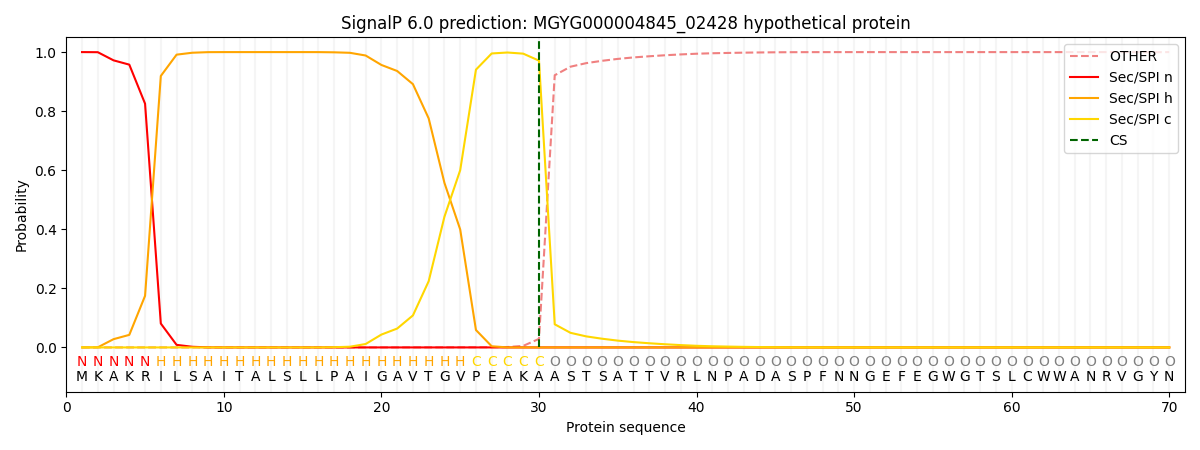
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000349 | 0.998886 | 0.000180 | 0.000229 | 0.000189 | 0.000169 |
