You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004848_00181
You are here: Home > Sequence: MGYG000004848_00181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004848_00181 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 218649; End: 221438 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 22 | 716 | 2.3e-73 | 0.726063829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.08e-25 | 28 | 476 | 12 | 457 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.82e-20 | 20 | 325 | 4 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.78e-13 | 100 | 451 | 113 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 1.15e-12 | 208 | 305 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 4.85e-10 | 31 | 168 | 4 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88596.1 | 0.0 | 24 | 929 | 22 | 928 |
| ALJ60397.1 | 0.0 | 24 | 929 | 22 | 928 |
| QDO68585.1 | 0.0 | 24 | 929 | 22 | 928 |
| SCD20869.1 | 0.0 | 24 | 916 | 27 | 916 |
| ARS42432.1 | 0.0 | 28 | 913 | 32 | 925 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 2.81e-24 | 29 | 452 | 39 | 464 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 2.81e-24 | 29 | 452 | 40 | 465 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 7SF2_A | 6.98e-18 | 100 | 451 | 90 | 454 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6ZJV_A | 2.87e-15 | 128 | 489 | 158 | 495 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
| 5T9A_A | 3.40e-15 | 96 | 433 | 85 | 439 | Crystalstructure of BuGH2Cwt [Bacteroides uniformis],5T9A_B Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9A_C Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9A_D Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9G_A Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_B Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_C Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_D Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 1.54e-23 | 29 | 452 | 40 | 465 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q6LL68 | 6.40e-12 | 128 | 484 | 148 | 519 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| A8GGN3 | 1.45e-11 | 27 | 450 | 53 | 482 | Beta-galactosidase OS=Serratia proteamaculans (strain 568) OX=399741 GN=lacZ PE=3 SV=1 |
| Q02603 | 9.70e-11 | 57 | 428 | 74 | 466 | Beta-galactosidase large subunit OS=Leuconostoc lactis OX=1246 GN=lacL PE=1 SV=1 |
| P26257 | 5.54e-10 | 27 | 450 | 3 | 411 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
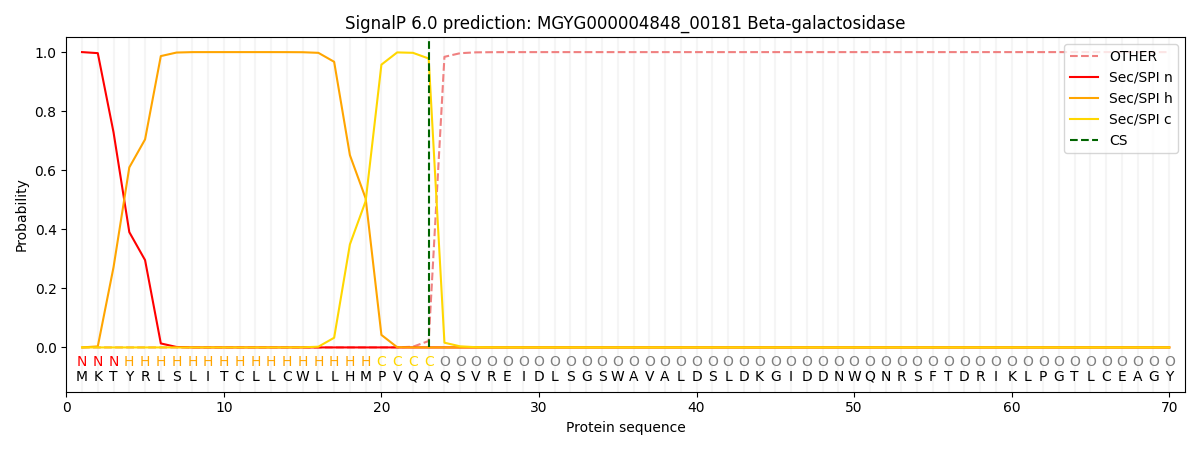
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000238 | 0.999083 | 0.000212 | 0.000157 | 0.000148 | 0.000141 |
