You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004850_00482
You are here: Home > Sequence: MGYG000004850_00482
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
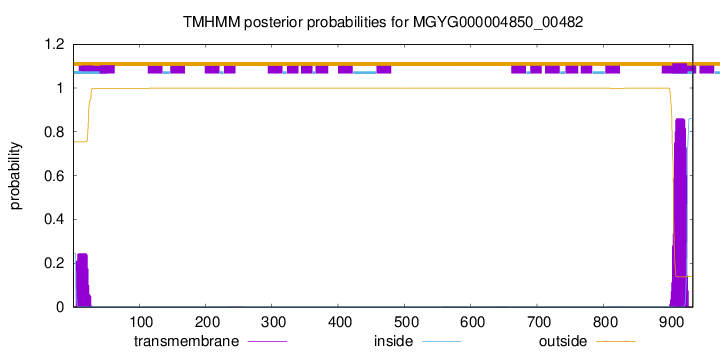
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; ; | |||||||||||
| CAZyme ID | MGYG000004850_00482 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18973; End: 21780 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 463 | 784 | 8.4e-63 | 0.8854166666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 8.42e-30 | 462 | 702 | 84 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 4.17e-29 | 462 | 762 | 6 | 253 | pectinesterase |
| pfam01095 | Pectinesterase | 1.14e-27 | 463 | 702 | 2 | 220 | Pectinesterase. |
| PLN02432 | PLN02432 | 3.00e-26 | 462 | 702 | 12 | 214 | putative pectinesterase |
| PLN02665 | PLN02665 | 3.73e-26 | 457 | 703 | 64 | 283 | pectinesterase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOG24296.1 | 1.69e-191 | 68 | 835 | 157 | 910 |
| QOL70560.1 | 5.11e-170 | 70 | 830 | 747 | 1492 |
| QLI95352.1 | 8.77e-166 | 47 | 831 | 721 | 1628 |
| AOP00060.1 | 5.72e-146 | 374 | 829 | 1 | 453 |
| AXF97721.1 | 1.24e-144 | 374 | 829 | 1 | 453 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 5.12e-26 | 469 | 820 | 15 | 335 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 9.11e-23 | 469 | 820 | 15 | 335 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 5C1E_A | 1.06e-22 | 463 | 703 | 11 | 224 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 1.43e-22 | 463 | 703 | 11 | 224 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 2NTB_A | 2.23e-22 | 469 | 820 | 15 | 335 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QK82 | 7.08e-22 | 463 | 703 | 39 | 252 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| P0C1A8 | 1.30e-21 | 469 | 820 | 39 | 359 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| P0C1A9 | 1.75e-21 | 469 | 820 | 39 | 359 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| Q5B7U0 | 2.27e-21 | 463 | 703 | 37 | 250 | Pectinesterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pmeA PE=1 SV=1 |
| Q9FM79 | 2.86e-21 | 462 | 709 | 81 | 307 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
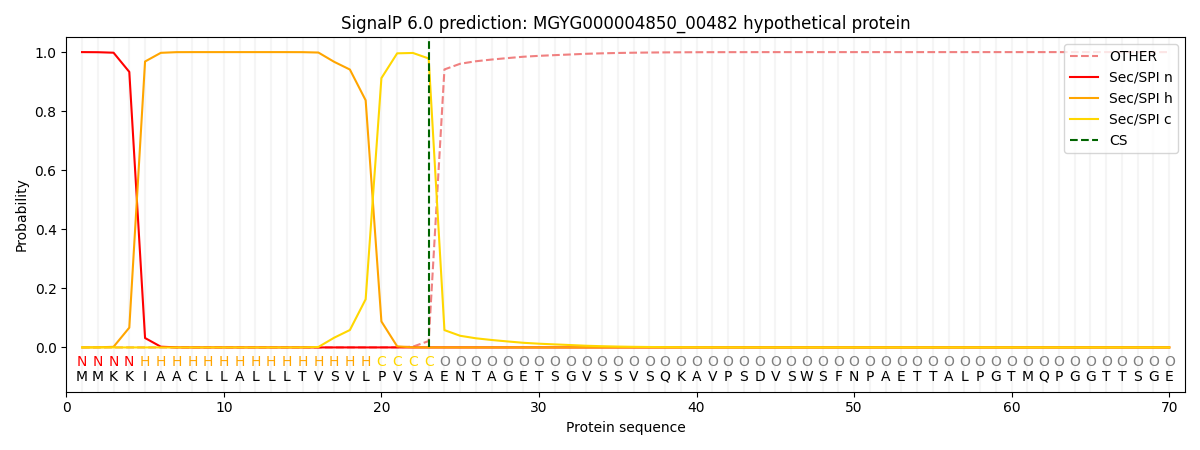
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000241 | 0.999049 | 0.000202 | 0.000170 | 0.000156 | 0.000140 |