You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004854_00212
You are here: Home > Sequence: MGYG000004854_00212
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; | |||||||||||
| CAZyme ID | MGYG000004854_00212 | |||||||||||
| CAZy Family | GH48 | |||||||||||
| CAZyme Description | Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 248579; End: 251068 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH48 | 42 | 742 | 4.8e-199 | 0.9950900163666121 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02011 | Glyco_hydro_48 | 0.0 | 42 | 745 | 1 | 620 | Glycosyl hydrolase family 48. Members of this family are endoglucanase EC:3.2.1.4 and exoglucanase EC:3.2.1.91 enzymes that cleave cellulose or related substrate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05724.1 | 0.0 | 1 | 776 | 1 | 797 |
| CBL17316.1 | 6.14e-291 | 1 | 823 | 1 | 799 |
| ADU23081.1 | 4.65e-231 | 1 | 748 | 1 | 779 |
| BAJ05814.1 | 4.65e-231 | 1 | 748 | 1 | 779 |
| CAS03459.1 | 6.58e-230 | 1 | 757 | 1 | 789 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D5D_A | 3.05e-158 | 42 | 745 | 20 | 642 | Structureof Caldicellulosiruptor danielii GH48 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 4XWL_A | 2.89e-156 | 39 | 750 | 45 | 676 | Catalyticdomain of Clostridium Cellulovorans Exgs [Clostridium cellulovorans],4XWM_A Complex structure of catalytic domain of Clostridium Cellulovorans Exgs and Cellobiose [Clostridium cellulovorans],4XWN_A Complex structure of catalytic domain of Clostridium Cellulovorans Exgs and Cellotetraose [Clostridium cellulovorans] |
| 4EL8_A | 2.09e-155 | 42 | 745 | 9 | 631 | Theunliganded structure of C.bescii CelA GH48 module [Caldicellulosiruptor bescii DSM 6725] |
| 4L0G_A | 4.15e-155 | 42 | 745 | 13 | 635 | CrystalStructure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725],4L6X_A Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725],4TXT_A Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725] |
| 5YJ6_A | 1.53e-151 | 41 | 747 | 12 | 636 | ChainA, Dockerin type I repeat-containing protein [Acetivibrio thermocellus DSM 1313] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH67 | 5.55e-150 | 23 | 777 | 20 | 693 | Cellulose 1,4-beta-cellobiosidase (reducing end) CelS OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celS PE=1 SV=1 |
| P0C2S5 | 5.55e-150 | 23 | 777 | 20 | 693 | Cellulose 1,4-beta-cellobiosidase (reducing end) CelS OS=Acetivibrio thermocellus OX=1515 GN=celS PE=1 SV=1 |
| P22534 | 3.30e-146 | 42 | 745 | 1117 | 1739 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P37698 | 2.17e-143 | 5 | 767 | 3 | 675 | Endoglucanase F OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCF PE=1 SV=2 |
| P50900 | 1.36e-142 | 36 | 745 | 34 | 655 | Exoglucanase-2 OS=Thermoclostridium stercorarium (strain ATCC 35414 / DSM 8532 / NCIMB 11754) OX=1121335 GN=celY PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
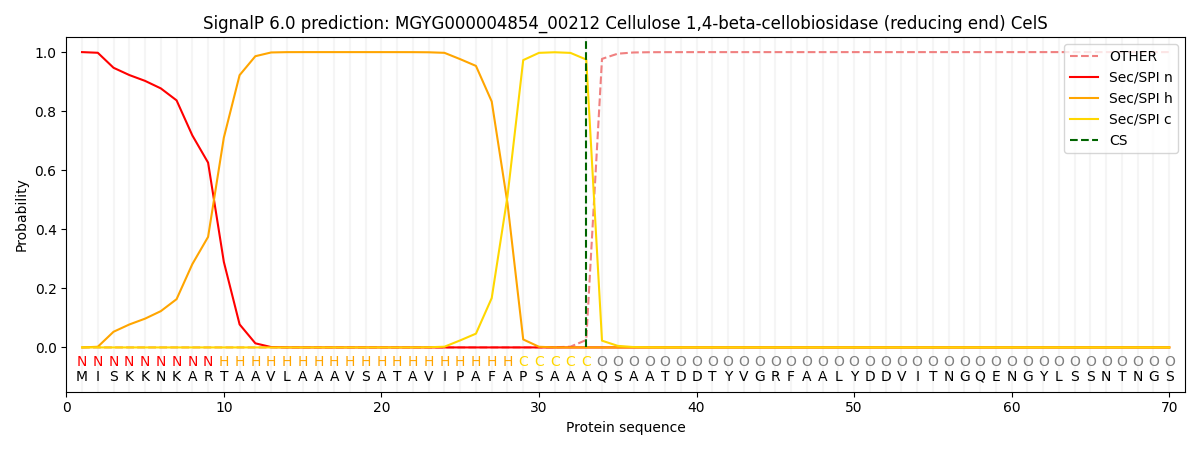
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000242 | 0.999011 | 0.000183 | 0.000209 | 0.000175 | 0.000147 |