You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004854_01676
You are here: Home > Sequence: MGYG000004854_01676
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
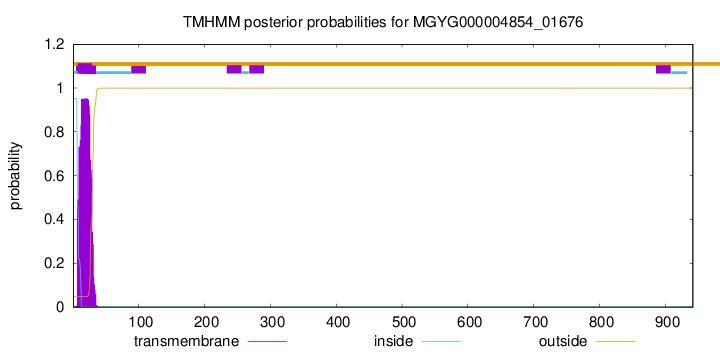
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; | |||||||||||
| CAZyme ID | MGYG000004854_01676 | |||||||||||
| CAZy Family | PL10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5873; End: 8701 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL10 | 75 | 343 | 1.2e-105 | 0.9756944444444444 |
| CE8 | 374 | 696 | 1.2e-56 | 0.90625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09492 | Pec_lyase | 4.18e-109 | 71 | 341 | 1 | 284 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| TIGR02474 | pec_lyase | 2.67e-105 | 71 | 341 | 1 | 285 | pectate lyase, PelA/Pel-15E family. Members of this family are isozymes of pectate lyase (EC 4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
| PLN02671 | PLN02671 | 3.46e-31 | 363 | 694 | 55 | 331 | pectinesterase |
| PLN02773 | PLN02773 | 1.01e-30 | 378 | 708 | 16 | 286 | pectinesterase |
| PLN02682 | PLN02682 | 1.19e-30 | 368 | 695 | 71 | 342 | pectinesterase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU21605.1 | 0.0 | 1 | 897 | 1 | 896 |
| QNU66057.1 | 5.23e-180 | 1 | 897 | 1 | 819 |
| ADL49995.1 | 8.46e-121 | 348 | 720 | 23 | 405 |
| ACL75601.1 | 2.24e-113 | 44 | 354 | 39 | 346 |
| AEY65183.1 | 4.68e-113 | 44 | 355 | 39 | 347 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R76_A | 1.05e-56 | 50 | 340 | 73 | 392 | ChainA, pectate lyase [Niveispirillum irakense] |
| 1GXM_A | 2.37e-56 | 60 | 341 | 33 | 318 | Family10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXM_B Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXN_A Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus] |
| 1GXO_A | 2.94e-55 | 60 | 341 | 33 | 318 | MutantD189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid [Cellvibrio japonicus] |
| 3UW0_A | 2.38e-09 | 370 | 693 | 35 | 342 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1QJV_A | 6.58e-09 | 464 | 716 | 77 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FM79 | 1.86e-18 | 381 | 694 | 94 | 352 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q2UBD9 | 2.08e-14 | 533 | 693 | 142 | 296 | Probable pectinesterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pmeA PE=3 SV=1 |
| B8NPS7 | 2.08e-14 | 533 | 693 | 142 | 296 | Probable pectinesterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pmeA PE=3 SV=1 |
| Q9ZQA3 | 6.02e-14 | 378 | 672 | 100 | 338 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| A1DBT4 | 1.21e-13 | 533 | 719 | 142 | 324 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
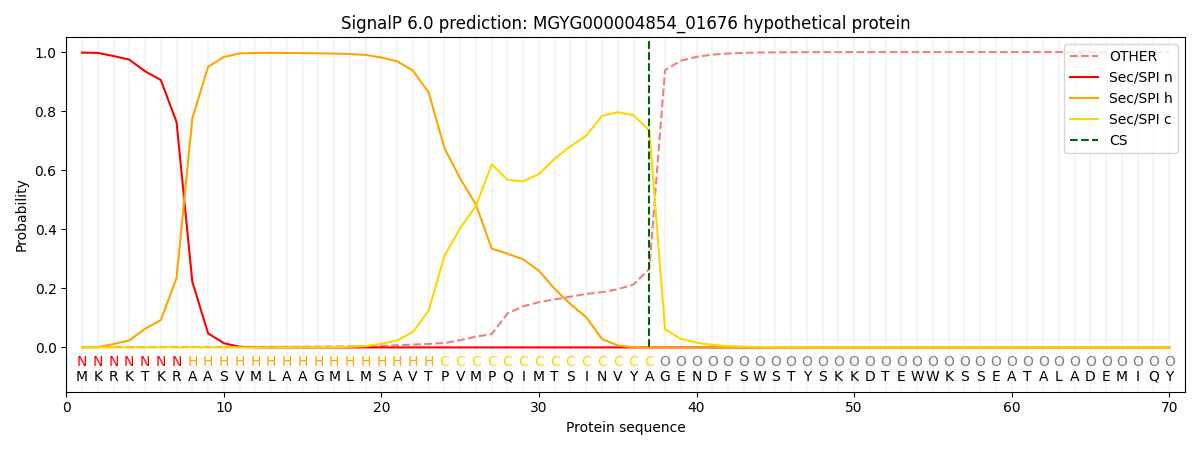
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002293 | 0.996095 | 0.000667 | 0.000505 | 0.000222 | 0.000186 |