You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004860_01904
You are here: Home > Sequence: MGYG000004860_01904
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
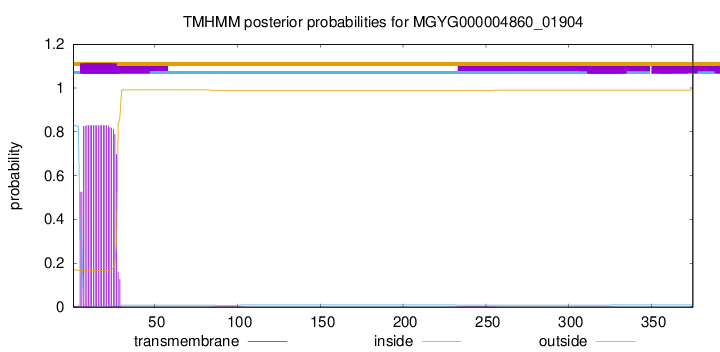
TMHMM annotations
Basic Information help
| Species | Levilactobacillus namurensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Levilactobacillus; Levilactobacillus namurensis | |||||||||||
| CAZyme ID | MGYG000004860_01904 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19105; End: 20232 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 55 | 372 | 1.8e-97 | 0.9906542056074766 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFR62464.1 | 1.42e-169 | 27 | 375 | 1 | 351 |
| QMU09191.1 | 3.06e-153 | 44 | 374 | 4 | 335 |
| AVK64526.1 | 1.57e-146 | 4 | 373 | 9 | 376 |
| SMS13688.1 | 3.96e-142 | 89 | 375 | 1 | 288 |
| AYG42170.1 | 7.23e-134 | 1 | 375 | 9 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 5.93e-21 | 52 | 372 | 23 | 344 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 1.20e-10 | 58 | 372 | 47 | 351 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
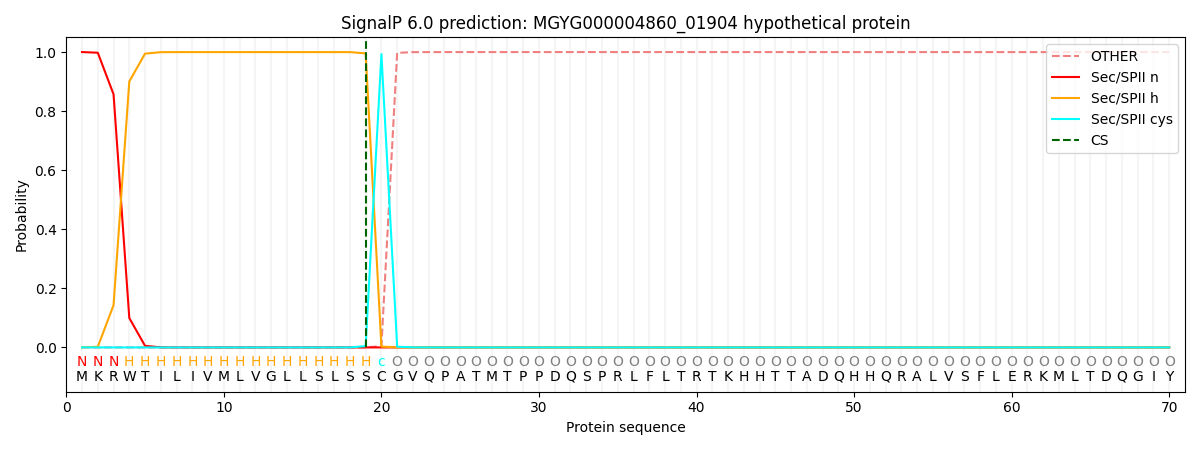
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000065 | 0.000000 | 0.000000 | 0.000000 |