You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004869_00304
You are here: Home > Sequence: MGYG000004869_00304
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactonifactor sp009677585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lactonifactor; Lactonifactor sp009677585 | |||||||||||
| CAZyme ID | MGYG000004869_00304 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91360; End: 93486 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM66 | 576 | 697 | 2.9e-17 | 0.8129032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03747 | ADP_ribosyl_GH | 5.29e-28 | 12 | 306 | 1 | 195 | ADP-ribosylglycohydrolase. This family includes enzymes that ADP-ribosylations, for example ADP-ribosylarginine hydrolase EC:3.2.2.19 cleaves ADP-ribose-L-arginine. The family also includes dinitrogenase reductase activating glycohydrolase. Most surprisingly the family also includes jellyfish crystallins, these proteins appear to have lost the presumed active site residues. |
| COG1397 | DraG | 2.48e-08 | 8 | 307 | 6 | 290 | ADP-ribosylglycohydrolase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYA15761.1 | 1.23e-163 | 2 | 708 | 3 | 702 |
| QTK81782.1 | 5.03e-158 | 2 | 700 | 3 | 688 |
| QRM46267.1 | 2.00e-157 | 2 | 700 | 3 | 688 |
| BCP52702.1 | 4.08e-150 | 2 | 706 | 7 | 704 |
| QYT00160.1 | 1.31e-141 | 2 | 708 | 4 | 704 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
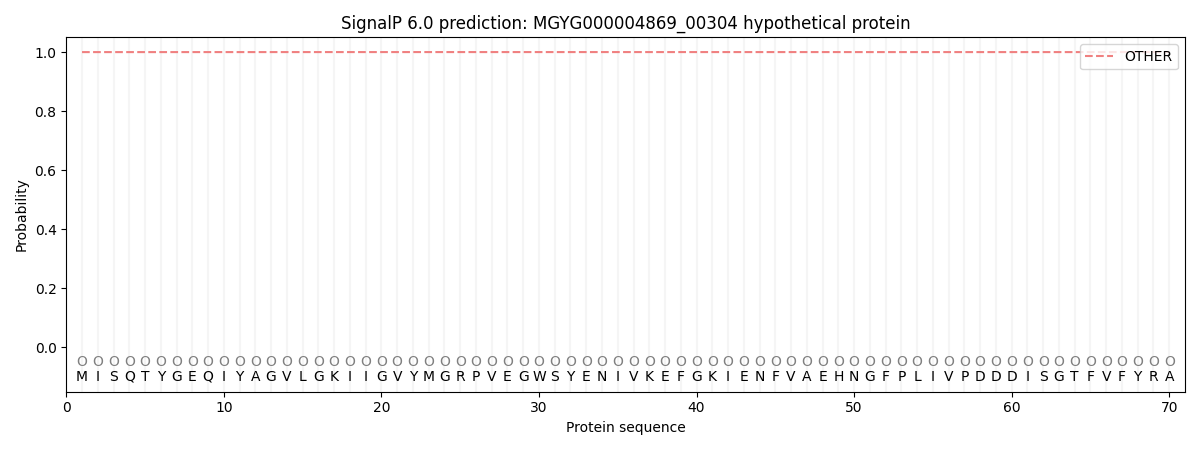
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000059 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
