You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004876_01502
You are here: Home > Sequence: MGYG000004876_01502
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000004876_01502 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 38909; End: 42010 Strand: - | |||||||||||
Full Sequence Download help
| MKKNILTGSL LLLCTALFSQ NFNEWRDPEV NEVNRAPMHT NYFAYQDEGS ALGRCKEASQ | 60 |
| NFMSLNGVWK FLWVKNADMR PVDFYKTGYD DKSWNDMQVP GLWALNGYGV PLYVNYGYPW | 120 |
| RNQFAGEPPT IPVEGNQVGS YRKEIIVPAD WREKEVFAHF GSVSSNIYLW VNGQFVGYSE | 180 |
| DSKLEAEFNL TKYLKPGKNL IAFQIFRWCD GSYLEDQDFL RLAGVGRDCY LYARNKKYIR | 240 |
| DIRVTPNLDA QYKDATLDVT VDIKGSGSVD LKLLDASGKE VVTTSVKGSG KVSTGMEVLN | 300 |
| PEKWSAETPV LYTLLATLKD KDRVLEVIPV NVGFRKIELR DSQIWVNGQP VLFKGVNRHE | 360 |
| MDPDNGYYVS PQRMIQDIRI MKAFNINAVR TSHYPNDNLW YDLCDRYGLY VVAEANVESH | 420 |
| GIGGHKTLAK NPLFAKAHLE RNQRNVQRSY NHPSIIFWSL GNEAGFGPNF EACYTWIKSE | 480 |
| DKTRAVQYEQ AGTNEFTDVY CPMYQDYEGN KKYCEGNIEK PLIQCEYTHA MGNSAGGFKE | 540 |
| YWDLIRKYPK YQGGFIWDFV DQSLRWKNKE GISFYAYGGD WSPYDASDNN FMDNGLISPD | 600 |
| RKPNPHMYEV GHFYQSVWAT PADLESGKIN VYNENFFRDL SAYYAEWELL ADGEVIRKGI | 660 |
| VDNLDVAPQQ TGIVRLGYAA DDICRDKEVL LNIFFKLRKA ETLLSAGHIV SRNQLVVRPY | 720 |
| KNLERSVDNV RGIKTRTDVP AIRDNDVNFL IIEGENFRLD FDKHDGYLCR YEMKGMQLVE | 780 |
| EGGKLKPNFW RAPTDNDLGA GLQMKYAAWK NTGIYLNKLE HKVADNLVQI DAEYTMNAVK | 840 |
| AKLYLTYVIN NNGAVKVTQR MFVDQSAKIS DLFRFGMQMR LPQELEQISY YGRGPGENYS | 900 |
| DRNHATFLGK YQQNVSSQFY SYIRPQENGT KTDIRWWKQT DRGGNGLKFV ADAPCSMSAL | 960 |
| HYTIESLDDG ERKDQRHSEF VPRVDYINLC IDKVQMGVGG VNSWGALPLE EYRLPYKDYE | 1020 |
| FSFFLLPLES DVE | 1033 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 56 | 908 | 2.5e-219 | 0.9680851063829787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 23 | 1025 | 15 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 23 | 1028 | 2 | 1000 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 4.43e-161 | 56 | 910 | 6 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 1.81e-104 | 753 | 1025 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 3.30e-100 | 337 | 617 | 1 | 300 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT51055.1 | 0.0 | 1 | 1032 | 1 | 1032 |
| QQA08535.1 | 0.0 | 1 | 1028 | 1 | 1031 |
| BCA52652.1 | 0.0 | 1 | 1028 | 1 | 1031 |
| QUT41248.1 | 0.0 | 1 | 1028 | 1 | 1031 |
| QMW85735.1 | 0.0 | 1 | 1028 | 1 | 1031 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 1.71e-248 | 24 | 1023 | 3 | 977 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 1.76e-248 | 24 | 1023 | 4 | 978 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 4.95e-214 | 20 | 1030 | 5 | 1003 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 7.01e-211 | 24 | 1030 | 6 | 999 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 1JZ7_A | 5.57e-170 | 24 | 1021 | 15 | 1016 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 9.64e-248 | 24 | 1023 | 4 | 978 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 3.83e-220 | 19 | 1028 | 14 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| P23989 | 2.51e-201 | 27 | 1028 | 13 | 1025 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
| Q9K9C6 | 2.57e-197 | 21 | 1023 | 6 | 1009 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q03WL0 | 1.63e-190 | 23 | 1023 | 11 | 1025 | Beta-galactosidase OS=Leuconostoc mesenteroides subsp. mesenteroides (strain ATCC 8293 / DSM 20343 / BCRC 11652 / CCM 1803 / JCM 6124 / NCDO 523 / NBRC 100496 / NCIMB 8023 / NCTC 12954 / NRRL B-1118 / 37Y) OX=203120 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
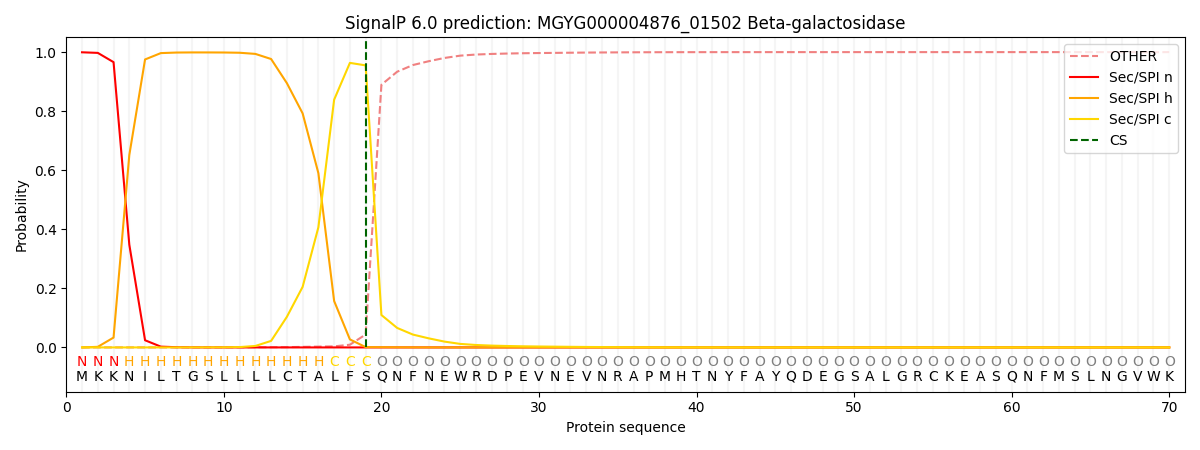
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000856 | 0.997464 | 0.001037 | 0.000211 | 0.000194 | 0.000196 |
