You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004891_02379
You are here: Home > Sequence: MGYG000004891_02379
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; KM106-2; | |||||||||||
| CAZyme ID | MGYG000004891_02379 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17553; End: 19712 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 6.00e-52 | 28 | 580 | 3 | 443 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 8.86e-33 | 88 | 243 | 1 | 155 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1287 | Stt3 | 8.68e-05 | 87 | 154 | 85 | 157 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB37647.1 | 1.33e-60 | 18 | 714 | 179 | 792 |
| QHU89858.1 | 5.20e-57 | 18 | 715 | 247 | 902 |
| QHU90800.1 | 5.78e-54 | 18 | 715 | 247 | 899 |
| QJU10517.1 | 5.19e-50 | 18 | 715 | 247 | 905 |
| QHU92512.1 | 1.67e-49 | 18 | 715 | 245 | 899 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 1.49e-08 | 88 | 252 | 87 | 256 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P37483 | 1.76e-96 | 29 | 719 | 4 | 671 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
| O34575 | 2.13e-93 | 29 | 699 | 5 | 664 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
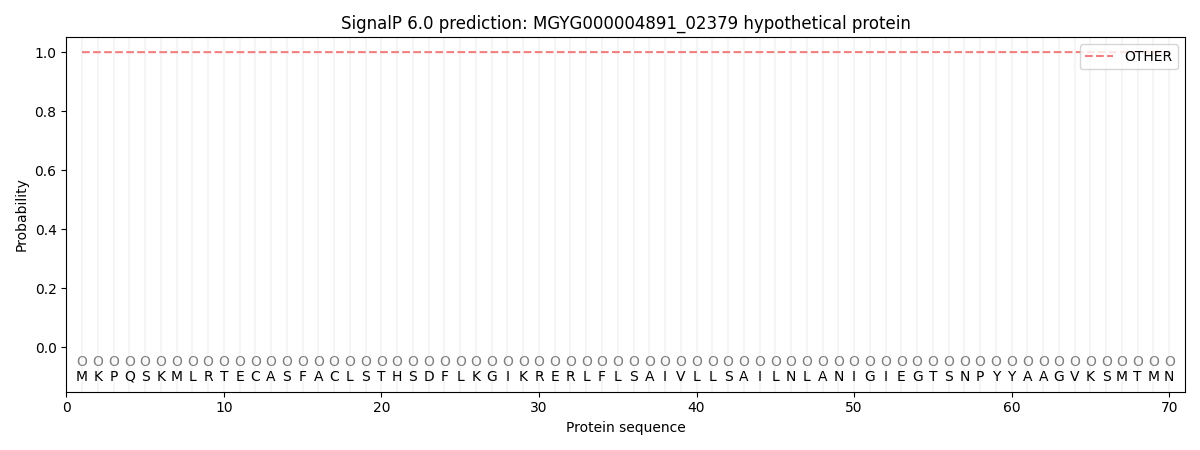
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999949 | 0.000050 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
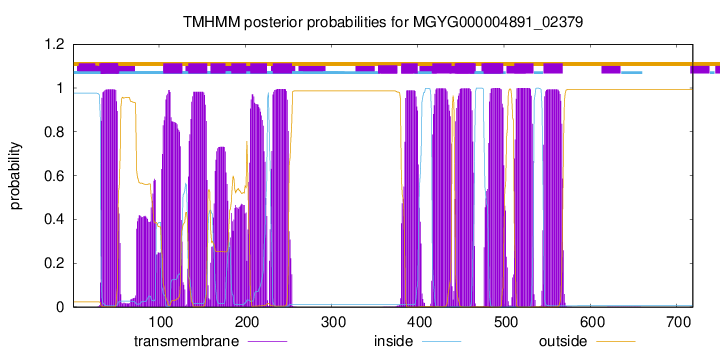
| start | end |
|---|---|
| 33 | 52 |
| 105 | 127 |
| 134 | 156 |
| 160 | 177 |
| 184 | 201 |
| 206 | 225 |
| 232 | 254 |
| 381 | 400 |
| 417 | 439 |
| 443 | 465 |
| 477 | 499 |
| 512 | 534 |
| 546 | 568 |
