You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004898_00686
You are here: Home > Sequence: MGYG000004898_00686
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Weissella confusa_B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Weissella; Weissella confusa_B | |||||||||||
| CAZyme ID | MGYG000004898_00686 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4293; End: 5630 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 1.63e-27 | 336 | 436 | 3 | 96 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 2.85e-25 | 95 | 431 | 140 | 466 | invasion associated endopeptidase. |
| COG0791 | Spr | 2.90e-18 | 246 | 428 | 12 | 179 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| cd00118 | LysM | 7.65e-14 | 154 | 198 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| NF033742 | NlpC_p60_RipB | 2.26e-13 | 335 | 437 | 90 | 198 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBZ04357.1 | 2.91e-245 | 1 | 445 | 1 | 445 |
| QIE78434.1 | 1.45e-194 | 1 | 444 | 1 | 438 |
| QYU57199.1 | 1.83e-190 | 1 | 444 | 1 | 438 |
| QBZ02383.1 | 3.99e-189 | 1 | 444 | 1 | 446 |
| QMU89441.1 | 8.99e-106 | 1 | 445 | 1 | 482 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B8C_A | 9.57e-23 | 322 | 442 | 30 | 141 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 3NE0_A | 4.18e-14 | 335 | 440 | 98 | 209 | Structureand functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation [Mycobacterium tuberculosis H37Rv] |
| 3PBC_A | 4.18e-14 | 335 | 440 | 98 | 209 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
| 4Q4T_A | 8.88e-14 | 335 | 440 | 356 | 467 | Structureof the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 [Mycobacterium tuberculosis H37Rv] |
| 2XIV_A | 1.76e-13 | 335 | 440 | 93 | 204 | Structureof Rv1477, hypothetical invasion protein of Mycobacterium tuberculosis [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13692 | 3.86e-20 | 322 | 431 | 404 | 505 | Protein P54 OS=Enterococcus faecium OX=1352 PE=3 SV=2 |
| P9WHU3 | 1.43e-13 | 311 | 428 | 264 | 370 | Probable endopeptidase Rv2190c OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv2190c PE=3 SV=1 |
| P67474 | 1.43e-13 | 311 | 428 | 264 | 370 | Probable endopeptidase Mb2213c OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2213C PE=3 SV=1 |
| P9WHU2 | 1.43e-13 | 311 | 428 | 264 | 370 | Probable endopeptidase MT2245 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2245 PE=3 SV=1 |
| Q01835 | 9.46e-13 | 321 | 431 | 397 | 496 | Probable endopeptidase p60 OS=Listeria grayi OX=1641 GN=iap PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
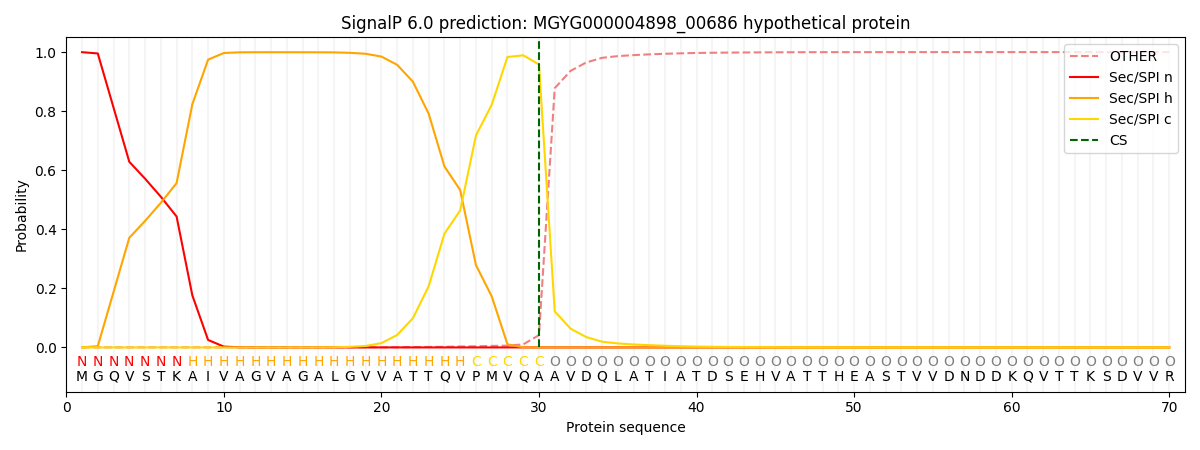
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001284 | 0.997766 | 0.000273 | 0.000243 | 0.000214 | 0.000196 |
