You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004899_02697
You are here: Home > Sequence: MGYG000004899_02697
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
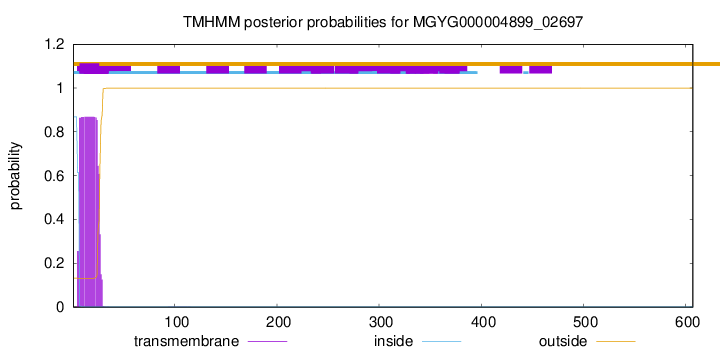
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900555635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900555635 | |||||||||||
| CAZyme ID | MGYG000004899_02697 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | O-GlcNAcase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18206; End: 20029 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 151 | 442 | 6.6e-115 | 0.9932203389830508 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 3.89e-178 | 151 | 441 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 3.18e-22 | 24 | 144 | 2 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam03648 | Glyco_hydro_67N | 0.006 | 99 | 125 | 87 | 117 | Glycosyl hydrolase family 67 N-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the N-terminal region of alpha-glucuronidase. The N-terminal domain forms a two-layer sandwich, each layer being formed by a beta sheet of five strands. A further two helices form part of the interface with the central, catalytic, module (pfam07488). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CUA20357.1 | 3.05e-300 | 1 | 607 | 1 | 608 |
| QCQ39276.1 | 3.05e-300 | 1 | 607 | 1 | 608 |
| AKA53424.1 | 3.05e-300 | 1 | 607 | 1 | 608 |
| CBW24319.1 | 3.05e-300 | 1 | 607 | 1 | 608 |
| ANQ61933.1 | 3.05e-300 | 1 | 607 | 1 | 608 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VVN_A | 1.58e-286 | 1 | 607 | 1 | 605 | BtGH84in complex with NH-Butylthiazoline [Bacteroides thetaiotaomicron VPI-5482],2VVN_B BtGH84 in complex with NH-Butylthiazoline [Bacteroides thetaiotaomicron VPI-5482],2VVS_A BtGH84 structure in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],2X0H_A BtGH84 Michaelis complex [Bacteroides thetaiotaomicron VPI-5482],2X0H_B BtGH84 Michaelis complex [Bacteroides thetaiotaomicron VPI-5482],4AIS_A A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482],4AIS_B A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482],7OU8_AAA Chain AAA, O-GlcNAcase BT_4395 [Bacteroides thetaiotaomicron VPI-5482],7OU8_BBB Chain BBB, O-GlcNAcase BT_4395 [Bacteroides thetaiotaomicron VPI-5482] |
| 2WZI_A | 9.04e-286 | 1 | 607 | 1 | 605 | BtGH84D243N in complex with 5F-oxazoline [Bacteroides thetaiotaomicron VPI-5482],2WZI_B BtGH84 D243N in complex with 5F-oxazoline [Bacteroides thetaiotaomicron VPI-5482] |
| 2WZH_A | 9.04e-286 | 1 | 607 | 1 | 605 | BtGH84D242N in complex with MeUMB-derived oxazoline [Bacteroides thetaiotaomicron VPI-5482],4AIU_A A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482] |
| 7K41_A | 2.98e-282 | 21 | 607 | 3 | 587 | ChainA, O-GlcNAcase BT_4395 [Escherichia coli K-12] |
| 2CHO_A | 3.81e-282 | 22 | 607 | 1 | 584 | Bacteroidesthetaiotaomicron hexosaminidase with O-GlcNAcase activity [Bacteroides thetaiotaomicron VPI-5482],2CHO_B Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q89ZI2 | 8.62e-286 | 1 | 607 | 1 | 605 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q0TR53 | 3.52e-112 | 25 | 547 | 45 | 583 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 3.52e-112 | 25 | 547 | 45 | 583 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| P26831 | 4.09e-62 | 26 | 519 | 41 | 577 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q8VIJ5 | 3.46e-42 | 152 | 410 | 63 | 331 | Protein O-GlcNAcase OS=Rattus norvegicus OX=10116 GN=Oga PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
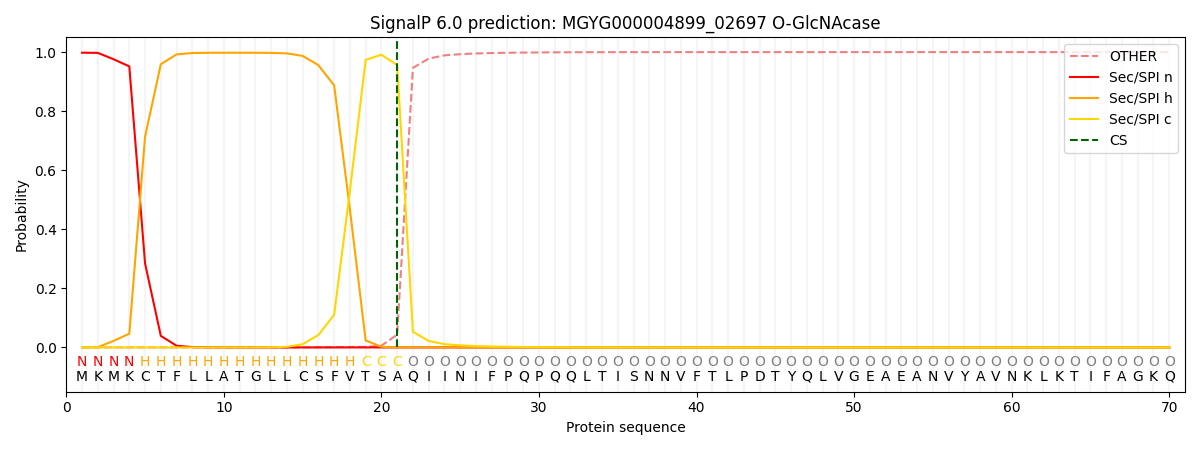
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000550 | 0.995937 | 0.002816 | 0.000233 | 0.000215 | 0.000210 |