You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004899_03363
You are here: Home > Sequence: MGYG000004899_03363
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
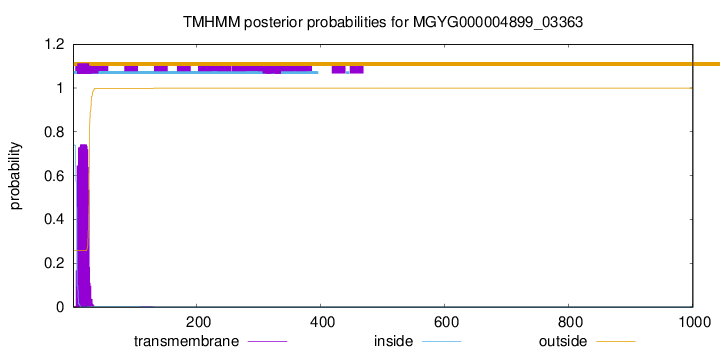
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900555635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900555635 | |||||||||||
| CAZyme ID | MGYG000004899_03363 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58111; End: 61116 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 42 | 615 | 7e-91 | 0.7837837837837838 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12708 | Pectate_lyase_3 | 3.36e-16 | 49 | 212 | 7 | 169 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam12708 | Pectate_lyase_3 | 6.20e-10 | 375 | 428 | 1 | 60 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG3386 | YvrE | 3.16e-09 | 714 | 994 | 15 | 277 | Sugar lactone lactonase YvrE [Carbohydrate transport and metabolism]. |
| pfam08450 | SGL | 6.31e-08 | 943 | 993 | 192 | 246 | SMP-30/Gluconolaconase/LRE-like region. This family describes a region that is found in proteins expressed by a variety of eukaryotic and prokaryotic species. These proteins include various enzymes, such as senescence marker protein 30 (SMP-30), gluconolactonase and luciferin-regenerating enzyme (LRE). SMP-30 is known to hydrolyze diisopropyl phosphorofluoridate in the liver, and has been noted as having sequence similarity, in the region described in this family, with PON1 and LRE. |
| cd14962 | NHL_like_6 | 2.15e-06 | 904 | 993 | 34 | 120 | Uncharacterized NHL-repeat domain in bacterial proteins. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT68913.1 | 0.0 | 1 | 1001 | 1 | 1000 |
| QUT62546.1 | 0.0 | 1 | 1001 | 1 | 1000 |
| QQA31096.1 | 0.0 | 1 | 1001 | 1 | 1000 |
| QBJ17508.1 | 0.0 | 1 | 1001 | 1 | 1000 |
| BBK89257.1 | 0.0 | 8 | 1001 | 2 | 994 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49426 | 2.54e-09 | 47 | 462 | 74 | 510 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
| Q37893 | 1.83e-06 | 376 | 428 | 104 | 156 | Pre-neck appendage protein OS=Bacillus phage B103 OX=10778 GN=12 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
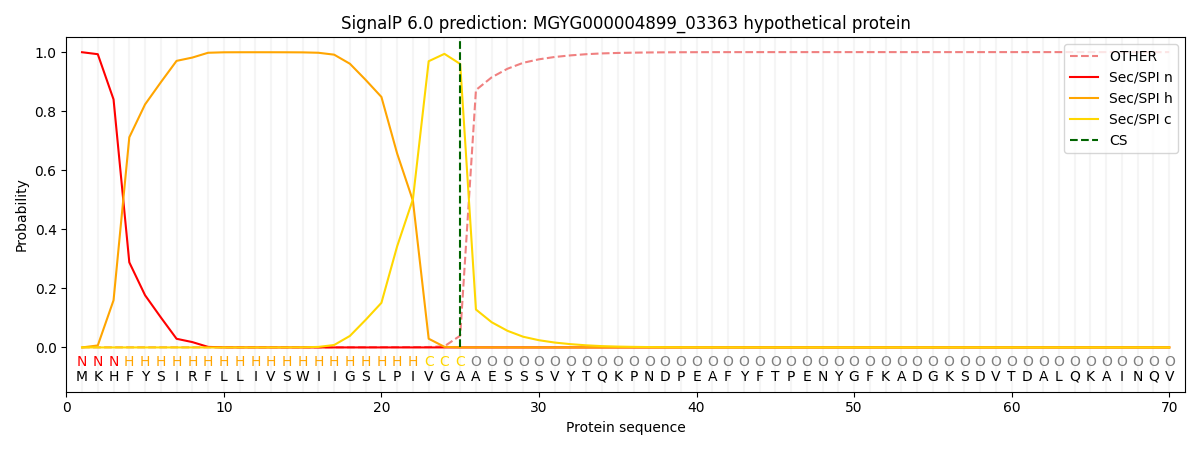
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001157 | 0.997980 | 0.000275 | 0.000212 | 0.000182 | 0.000174 |