

| Basic Information | |
|---|---|
| Species | Panicum virgatum |
| Cazyme ID | Pavirv00047982m |
| Family | AA1 |
| Protein Properties | Length: 504 Molecular Weight: 53787.6 Isoelectric Point: 6.5727 |
| Chromosome | Chromosome/Scaffold: 0159113 Start: 882 End: 2684 |
| Description | laccase 7 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 16 | 498 | 0 |
| SITAVDGVPGPVIEASEGDTVVVHVINDSPYDVTIHWHGVFQRGTPWADGPSMVTQCPIRQGSRYTYRFSVAGQEGTLWWHAHSSFLRATVHGALIIRPS AGAYPFPKPDGEAVVLLGEWWDADIALLERQAFLSGTQIPSADAYTINGKPNRTEKFSVRGNSTYLLRIINAALNTAFFFKVAGHTFTVVAADASYTTPY ETDVIVTAPGQTVNALMAADATPGRYYMAISSYQSAMPLRLESFNGDVTTAVVEYVGAAAAAPAGQQAAPPALPEMPEPTDTTTANRFYTGMTARVLPGR PTVPLAVDTRMLVTVGLGLSSCQPEQTSCNRSAPVVVASMNNESFALPTTTSLLDARYGGSAPHHGVSTSDFPDRPPAAFDYTNATGLVPAALSWTGRPT TKVRALRYNATVEVVLQNTALVARESHPMHLHGFDFFVLAQGFGNYDAGAASERLNLVNPQQRNTVAVPTGGWAVIRFVADNP | |||
| Full Sequence |
|---|
| Protein Sequence Length: 504 Download |
| VGTLSWPRIC QPGNLSITAV DGVPGPVIEA SEGDTVVVHV INDSPYDVTI HWHGVFQRGT 60 PWADGPSMVT QCPIRQGSRY TYRFSVAGQE GTLWWHAHSS FLRATVHGAL IIRPSAGAYP 120 FPKPDGEAVV LLGEWWDADI ALLERQAFLS GTQIPSADAY TINGKPNRTE KFSVRGNSTY 180 LLRIINAALN TAFFFKVAGH TFTVVAADAS YTTPYETDVI VTAPGQTVNA LMAADATPGR 240 YYMAISSYQS AMPLRLESFN GDVTTAVVEY VGAAAAAPAG QQAAPPALPE MPEPTDTTTA 300 NRFYTGMTAR VLPGRPTVPL AVDTRMLVTV GLGLSSCQPE QTSCNRSAPV VVASMNNESF 360 ALPTTTSLLD ARYGGSAPHH GVSTSDFPDR PPAAFDYTNA TGLVPAALSW TGRPTTKVRA 420 LRYNATVEVV LQNTALVARE SHPMHLHGFD FFVLAQGFGN YDAGAASERL NLVNPQQRNT 480 VAVPTGGWAV IRFVADNPAL HGR* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam00394 | Cu-oxidase | 2.0e-32 | 126 | 270 | 150 | + Multicopper oxidase. Many of the proteins in this family contain multiple similar copies of this plastocyanin-like domain. | ||
| pfam07732 | Cu-oxidase_3 | 2.0e-42 | 16 | 117 | 105 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| PLN02191 | PLN02191 | 4.0e-52 | 6 | 500 | 538 | + L-ascorbate oxidase | ||
| TIGR03388 | ascorbase | 4.0e-66 | 14 | 498 | 517 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03389 | laccase | 7.0e-176 | 16 | 498 | 494 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | EEE55676.1 | 0 | 1 | 500 | 11 | 519 | hypothetical protein OsJ_04090 [Oryza sativa Japonica Group] |
| RefSeq | NP_001066518.1 | 0 | 1 | 500 | 31 | 535 | Os12g0259800 [Oryza sativa (japonica cultivar-group)] |
| Swiss-Prot | Q5N7A3 | 0 | 1 | 500 | 31 | 539 | LAC6_ORYSJ RecName: Full=Laccase-6; AltName: Full=Benzenediol:oxygen oxidoreductase 6; AltName: Full=Urishiol oxidase 6; AltName: Full=Diphenol oxidase 6; Flags: Precursor |
| RefSeq | XP_002442097.1 | 0 | 1 | 500 | 34 | 535 | hypothetical protein SORBIDRAFT_08g011530 [Sorghum bicolor] |
| RefSeq | XP_002456639.1 | 0 | 1 | 500 | 30 | 526 | hypothetical protein SORBIDRAFT_03g039960 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 0 | 17 | 500 | 24 | 502 | X Chain X, Crystal Structure Of The Full-Length Autotransporter Esta From Pseudomonas Aeruginosa |
| PDB | 1asq_A | 0 | 17 | 500 | 24 | 502 | X Chain X, Crystal Structure Of The Full-Length Autotransporter Esta From Pseudomonas Aeruginosa |
| PDB | 1asp_B | 0 | 17 | 500 | 24 | 502 | X Chain X, Crystal Structure Of The Full-Length Autotransporter Esta From Pseudomonas Aeruginosa |
| PDB | 1asp_A | 0 | 17 | 500 | 24 | 502 | X Chain X, Crystal Structure Of The Full-Length Autotransporter Esta From Pseudomonas Aeruginosa |
| PDB | 1aso_B | 0 | 17 | 500 | 24 | 502 | X Chain X, Crystal Structure Of The Full-Length Autotransporter Esta From Pseudomonas Aeruginosa |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
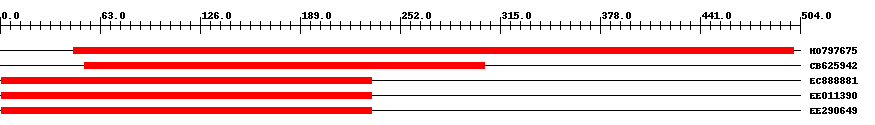 | ||||
| Hit | Length | Start | End | EValue |
| HO797675 | 469 | 46 | 500 | 0 |
| CB625942 | 266 | 53 | 305 | 0 |
| EC888881 | 246 | 1 | 234 | 0 |
| EE011390 | 246 | 1 | 234 | 0 |
| EE290649 | 246 | 1 | 234 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |