

| Basic Information | |
|---|---|
| Species | Panicum virgatum |
| Cazyme ID | Pavirv00062920m |
| Family | AA1 |
| Protein Properties | Length: 522 Molecular Weight: 56466.4 Isoelectric Point: 6.1868 |
| Chromosome | Chromosome/Scaffold: 001848 Start: 5479 End: 7366 |
| Description | laccase 7 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 3 | 508 | 0 |
| ISQLCMNSVVYAVNQQLPGPTVEVSEGDTLVVHVVNASPYPISIHWHGIFQLQSGWADGANMITQCPIQPFAKFTYVFSVTGQEGTLWWHAHASMLRATI YGALIIKPRSGYPFPPPHAEIPILLGEWWNRNVDDVENDGLLTGLGPAMSDAFTINGSPGDQAPCGGAGMFQVEVESGKTYLLRIINAAVNAELFFRVAG HAFTVVAADASYINPHPTDVIVVAPGQTVDALMDAAAAPGRYYMAARAFESKTVPAPPPFDTTTATAVLRYKGVPDYDSSAPAAMPALPPYTDVVTAARF YWSLIGLVRPGDPVIPRTVDYSLVVAFGVEQAPCAPDQTRCQGFSVLASMNRHSFRFPEKVSLLEALFRGVPNVYSEDFPGWPAAPATTKATSVRKVNFN DVVEVVLQNEGYSRALGTENHPIHLHGFNFFVLAQGLGRFDPRMRSTYNLENPQVRNTVAVPAGGWAVIRFTANNPGMWFMHCHLDAHLPAGLAMVFEVL NGPAPN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 522 Download |
| MGISQLCMNS VVYAVNQQLP GPTVEVSEGD TLVVHVVNAS PYPISIHWHG IFQLQSGWAD 60 GANMITQCPI QPFAKFTYVF SVTGQEGTLW WHAHASMLRA TIYGALIIKP RSGYPFPPPH 120 AEIPILLGEW WNRNVDDVEN DGLLTGLGPA MSDAFTINGS PGDQAPCGGA GMFQVEVESG 180 KTYLLRIINA AVNAELFFRV AGHAFTVVAA DASYINPHPT DVIVVAPGQT VDALMDAAAA 240 PGRYYMAARA FESKTVPAPP PFDTTTATAV LRYKGVPDYD SSAPAAMPAL PPYTDVVTAA 300 RFYWSLIGLV RPGDPVIPRT VDYSLVVAFG VEQAPCAPDQ TRCQGFSVLA SMNRHSFRFP 360 EKVSLLEALF RGVPNVYSED FPGWPAAPAT TKATSVRKVN FNDVVEVVLQ NEGYSRALGT 420 ENHPIHLHGF NFFVLAQGLG RFDPRMRSTY NLENPQVRNT VAVPAGGWAV IRFTANNPGM 480 WFMHCHLDAH LPAGLAMVFE VLNGPAPNLL PPPPADYPKC H* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam07731 | Cu-oxidase_2 | 1.0e-37 | 389 | 501 | 113 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| PLN02168 | PLN02168 | 2.0e-39 | 6 | 481 | 489 | + copper ion binding / pectinesterase | ||
| pfam07732 | Cu-oxidase_3 | 6.0e-44 | 3 | 113 | 113 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| TIGR03388 | ascorbase | 2.0e-73 | 7 | 500 | 533 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03389 | laccase | 3.0e-179 | 3 | 520 | 538 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| DDBJ | BAD82649.1 | 0 | 3 | 479 | 35 | 511 | putative laccase LAC6-8 [Oryza sativa Japonica Group] |
| GenBank | EEC71812.1 | 0 | 3 | 521 | 35 | 553 | hypothetical protein OsI_04450 [Oryza sativa Indica Group] |
| RefSeq | NP_001044816.1 | 0 | 3 | 479 | 35 | 511 | Os01g0850800 [Oryza sativa (japonica cultivar-group)] |
| Swiss-Prot | Q0JHP8 | 0 | 3 | 521 | 35 | 553 | LAC8_ORYSJ RecName: Full=Laccase-8; AltName: Full=Benzenediol:oxygen oxidoreductase 8; AltName: Full=Urishiol oxidase 8; AltName: Full=Diphenol oxidase 8; Flags: Precursor |
| RefSeq | XP_002456641.1 | 0 | 1 | 521 | 36 | 557 | hypothetical protein SORBIDRAFT_03g039980 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 0 | 7 | 499 | 19 | 521 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1asq_A | 0 | 7 | 499 | 19 | 521 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1asp_B | 0 | 7 | 499 | 19 | 521 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1asp_A | 0 | 7 | 499 | 19 | 521 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1aso_B | 0 | 7 | 499 | 19 | 521 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
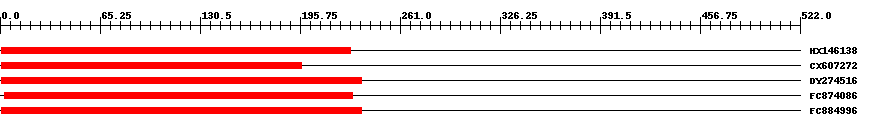 | ||||
| Hit | Length | Start | End | EValue |
| HX146138 | 232 | 1 | 229 | 0 |
| CX607272 | 200 | 1 | 197 | 0 |
| DY274516 | 238 | 1 | 236 | 0 |
| FC874086 | 230 | 3 | 230 | 0 |
| FC884996 | 238 | 1 | 236 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |